-
PDF
- Split View
-
Views
-
Cite
Cite
Evropi Theodoratou, Zahra Montazeri, Steven Hawken, Genevieve CdL Allum, Jacintha Gong, Valerie Tait, Iva Kirac, Mahmood Tazari, Susan M Farrington, Alex Demarsh, Lina Zgaga, Denise Landry, Helen E Benson, Stephanie H Read, Igor Rudan, Albert Tenesa, Malcolm G Dunlop, Harry Campbell, Julian Little, Systematic Meta-Analyses and Field Synopsis of Genetic Association Studies in Colorectal Cancer, JNCI: Journal of the National Cancer Institute, Volume 104, Issue 19, 3 October 2012, Pages 1433–1457, https://doi.org/10.1093/jnci/djs369
Close - Share Icon Share
Abstract
Colorectal cancer is a major global public health problem, with approximately 950 000 patients newly diagnosed each year. We report the first comprehensive field synopsis and creation of a parallel publicly available and regularly updated database (CRCgene) that catalogs all genetic association studies on colorectal cancer (http://www.chs.med.ed.ac.uk/CRCgene/).
We performed two independent systematic reviews, reviewing 10 145 titles, then collated and extracted data from 635 publications reporting on 445 polymorphisms in 110 different genes. We carried out meta-analyses to derive summary effect estimates for 92 polymorphisms in 64 different genes. For assessing the credibility of associations, we applied the Venice criteria and the Bayesian False Discovery Probability (BFDP) test.
We consider 16 independent variants at 13 loci (MUTYH, MTHFR, SMAD7, and common variants tagging the loci 8q24, 8q23.3, 11q23.1, 14q22.2, 1q41, 20p12.3, 20q13.33, 3q26.2, 16q22.1, and 19q13.1) to have the most highly credible associations with colorectal cancer, with all variants except those in MUTYH and 19q13.1 reaching genome-wide statistical significance in at least one meta-analysis model. We identified less-credible (higher heterogeneity, lower statistical power, BFDP >0.2) associations with 23 more variants at 22 loci. The meta-analyses of a further 20 variants for which associations have previously been reported found no evidence to support these as true associations.
The CRCgene database provides the context for genetic association data to be interpreted appropriately and helps inform future research direction.
Colorectal cancer is a major global public health problem, with approximately 950 000 patients newly diagnosed each year (www.who.int; International Agency for Research on Cancer GLOBOCAN). The risk of developing colorectal cancer increases steeply with age and its incidence is rising in many industrialized countries as life expectancy and the number of elderly people increase. Incidence is also rising in many developing countries as diet and lifestyle change and increasingly resemble those in industrialized countries. Overall, 5-year survival of colorectal cancer remains at best only approximately 50%. Because of the rising disease burden, health care costs, including costs for diagnosis and treatment, are substantial.
Approximately one-third of variance in colorectal cancer is attributed to inherited genetic factors (1) and disease risk is increased by two- to fourfold for first-degree relatives of patients. Excess risk of familial cancer can be accounted for by a combination of rare high-penetrance mutations and large numbers of common genetic variants, each of which confers a small risk. These variants combine to confer a range of susceptibilities in the population (2).
The associations between genetic variants and human diseases described so far were highly dependent on the study designs used to identify them. Highly penetrant mutations with large effects have been identified in a number of genes responsible for heritable colorectal cancer susceptibility syndromes (eg, DNA mismatch repair genes, APC, SMAD4, LKB1/STK11, MUTYH, and a linkage region on 15q) and these mutations account for about 3% of incident colorectal cancers in industrialized countries. Identification of further high-penetrance loci has proven intractable to genetic linkage approaches, probably because of locus heterogeneity, and confounding, because of the segregation of multiple alleles in collected families. Association studies conducted in general population samples using common genetic markers have typically identified variants with very small effects. To date, genome-wide association studies (GWASs) have reported 14 common genetic variants that influence the risk of colorectal cancer and account for approximately 6% of the excess familial risk (2–10). Future resequencing studies are expected to identify rarer variants (eg, with 0.05%–5% prevalence) with intermediate, or perhaps even large, effects (2). GWASs of structural variations will likely identify deletions, amplifications, and other copy-number variations that may also influence risk of colorectal cancer.
For this report, we have undertaken a comprehensive review of genetic factors that appear to be associated with colorectal cancer by using previously published guidelines for the assessment of cumulative evidence on genetic association studies (11,12) and following a format similar to previous overview meta-analyses (13–15). We have cataloged all genetic association studies published in this field and conducted meta-analyses of variants with genotypes available in four or more independent case-control studies; any variants that had been typed in two large GWASs were also included in the meta-analyses. The results of the search strategy and meta-analyses are publicly available on a regularly updated Internet database (CRCgene). This represents the first attempt to systematically capture all published genetic association data for colorectal cancer and conduct a meta-analysis.
The core aim is to provide an up-to-date systematic review of the state of the art across the field of colorectal cancer genetics for the research community. We conducted a critical review of all published “candidate gene” study data, incorporating relevant candidate gene data from the GWASs available to us, and then performed meta-analyses of these data. This approach enabled us to summarize available evidence from larger sample sizes, thus gaining greater precision in odds ratio estimates. We have presented these data within a defined statistical and causal inference framework to aid correct interpretation of data (16).
The broader medium-term aim of this work is to identify genetic variants for which there is robust evidence of influence on risk of colorectal cancer. This will help to inform future research efforts and to identify variants that can serve as a basis for providing risk estimates for population groups. It will also provide new insights into the fundamental biological mechanisms involved in colorectal carcinogenesis.
Methods
Literature Search and Data Collection
The first step was to undertake a comprehensive systematic literature review of all current published data on genetics and colorectal cancer. To identify gene association studies aimed at risk of colorectal cancer, we used the Medline database via the Ovid gateway and the search terms comprising medical subject headings (MeSH) and keywords relating to colorectal neoplasms, the MeSH heading “genetic predisposition to disease,” and the keywords “gene$” and “associate$” were applied to terms in the entire article. We cross-checked these 10 125 findings against those listed in the HuGENet phenopedia (17). Of the 10 145 articles thus identified, first we screened the abstracts for eligibility, and then, if necessary, the full texts. We used the following inclusion and exclusion criteria. The article must have evaluated the association between a polymorphic genetic variant [one with a minor allele frequency (MAF) ≥0.01 in the general population based on the data on the reference panel of the 1000 GenomesTable 1; (18)] and sporadic colorectal cancer. Studies that examined associations with only premalignant conditions such as adenomas, polyps, or dysplastic tissue were not included. In addition, studies of hereditary colorectal cancer syndromes, such as familial adenomatous polyposis, hereditary nonpolyposis colorectal cancer, juvenile polyposis syndrome, and Gardner’s syndrome were excluded because our focus was on sporadic colorectal cancer. All studies needed to relate to human participants; any study that was concerned solely with investigating the progression or histological phenotype of colorectal cancer was excluded. Case–control studies and appropriate cohort and GWASs were included. The study had to be published in English in a peer review journal before June 30, 2010. For variants that were identified through the GWAS, the search was repeated and extended until March 31, 2011. Any research that had only been reported in abstracts (eg, presented in scientific conferences yet to be fully published) was excluded. Nine family-based studies were also excluded. We generated a list with all variants to be summarized using meta-analysis and compared it with a list of variants that were included in two GWASs, from Scotland and Canada, for which we had access to individual-level data. The Scottish study comprised patients aged 16–79 years who were diagnosed with colorectal cancer throughout Scotland during the period 1999–2006 (6,19). Potentially eligible population-based control subjects were selected through the Community Health Index, a national register of all individuals registered with a general practitioner in Scotland, and individually matched with patients on age (± 2 years), sex, and area of residence. The Canadian study was based on patients aged 20–74 years who were diagnosed during the period July 1, 1997, to June 30, 2000, who were identified and recruited from the population-based Ontario Cancer Registry (3,20). Random digit dialing was used to select population-based control subjects who were frequency-matched with patients on sex and 5-year age group. In both of these studies, DNA from blood was genotyped using multiple platforms; here, we included data obtained in both studies from the Illumina HumanHap300 and HumanHap240S arrays on the Infinium platform. If a variant was found to be included in either of these GWASs, then genotype counts were included in the meta-analysis of this variant. For the other studies, we contacted the authors of studies with missing data, with an approximate successful response rate of 0.2.
Data Entry, Management, and Abstraction
Once the search was completed, the references of the articles in the search were entered into a web-based database, “RefWorks” (http://www.refworks.com/), thus freezing the search at that point in time. Their abstracts and/or full texts were screened to assess their eligibility for inclusion in the field synopsis. Review articles and meta-analyses on genetic associations of colorectal cancer were also kept alongside so that the references they used could be screened for eligibility if they had been missed in the Medline search.
Data from all studies that met the final inclusion and exclusion criteria were abstracted into two standardized tables. The first of these listed study characteristics and the second table listed allele and genotype frequencies. We abstracted key variables with regard to the study identifiers and context, study design and limitations, intervention specifics, and outcome effects.
Statistical Analysis
Statistical analysis was conducted using Intercooled STATA, version 11.0 (21). Meta-analysis was performed for all variants with case-control data available from four or more independent samples. We obtained summary crude odds ratios (ORs) and 95% confidence intervals (95% CI) for two additive models (variant [var]/wild-type [wt] vs wt/wt; and var/var vs wt/wt), one recessive model (var/var vs var/wt and wt/wt), and one dominant model (var/var and var/wt vs wt/wt). We applied either the fixed-effect model (Mantel–Haenszel method) or, in case of heterogeneity, the random-effect model (DerSimonian–Laird method). Between-study heterogeneity was quantified by calculating the Q statistic, with a P value less than .05 being the threshold. We also calculated the I2 heterogeneity metric and its 95% confidence interval (CI). Although sometimes we summarized studies that were very heterogeneous, we recognized that because of the variation in study methods and outcome definitions, the meta-estimates should be interpreted cautiously. To assess any small-study effects, we performed funnel plot analyses and tested for statistical significance using the Harbord modification of the Egger test, implemented in STATA (http://ideas.repec.org/a/tsj/stataj/v9y2009i2p197-210.html). A negative result from small study–effects testing does not entirely exclude publication bias. In addition, the test for small-study effects may be underpowered if there are approximately 10 or fewer studies and it may be inappropriate in the presence of large heterogeneity (22). We also estimated the power that each meta-analysis had to detect a statistically significant effect by using the Power and Sample Size Program (23) and specifying α = .05 as the level of statistical significance and the effect sizes and allele frequencies estimated from the meta-analyses (an integral component of the BFDP analysis).
The relative risk of colorectal cancer in a sibling that was attributable to a given single-nucleotide polymorphism (SNP) was calculated using the following formula (24,25):
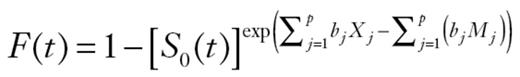
where p is the population frequency of the referent allele, q = 1– p, and r1 and r2 are the relative risks (estimated as ORs from the meta- analyses) for heterozygotes and variant homozygotes, relative to wild-type homozygotes. Assuming a multiplicative interaction, we calculated the proportion of the familial risk attributable to an SNP as log(λ*)/log(λ0), where λ0 is the overall familial relative risk estimated from epidemiological studies, assumed to be 2.2. Although family studies are normally used to estimate the relative risk for siblings, this formula can be used in population-based studies (24,25).
Finally, we repeated the analysis by excluding any studies that were conducted in a nonwhite population (with white populations defined as Europeans, North Americans, and Australians) for the SNPs that were found to be statistically significantly associated with risk of colorectal cancer in any of the genetic models at a threshold level of α = .05.
Credibility of Genetic Association
To assess the credibility of genetic associations, we considered the BFDP (26) and the Venice criteria (11,12). The BFDP assesses the noteworthiness of an observed association. For variants that were found to be statistically significantly associated with risk of colorectal cancer in any of the genetic models (at P < .05), the BFDP was estimated using the Excel Calculation Spreadsheet (http://faculty.washington.edu/jonno/cv.html). The BFDP threshold for noteworthiness was set up to be equal to 0.20, based on the assumption that a false discovery would be four times more costly than a false nondiscovery. We chose to calculate BFDP values for two levels of prior probabilities: at a medium or low prior level (0.05 to 10–3) that would be close to what would be expected for a candidate gene; and at a very low prior level (10–4 to 10–6) that would be close to what would be expected for a random SNP.
With regard to the Venice criteria, we operationalized the criterion of volume of evidence on the basis of statistical power to detect an association of the desired magnitude: A, 80% or more; B, 50%–79%; or C, less than 50%. For replication, we used the I² criterion proposed by Ioannidis et al. (11). For protection against bias, we considered that complete reporting was problematic. The phenotype definition was addressed by our inclusion criterion ––namely, that case subjects would have newly incident colorectal cancer. In general, genotyping error rates are low (27), and the criterion of replication across studies in part addressed potential concern about variation in genotyping quality between studies; some genotyping issues are discussed in relation to specific findings below. Whereas population stratification may impact gene discovery (28,29), the effect on the magnitude of association in general appears to be small (30,31).
We classified the genetic associations in three categories according to the findings after the BFDP analysis and the application of the Venice criteria. Associations were classified as positive if they fulfilled the following criteria: 1) they were statistically significant (at P < .05) in at least two of the genetic models; 2) they had a BFDP less than 0.20 (at least when P < .05); 3) they had a statistical power greater than 80%; and 4) they had an I2 less than 50%. Associations were classified as less-credible positives if they were statistically significant (P < .05) in at least one of the genetic models, their BFDP was greater than 0.20, and their statistical power was between 50% and 79% (I2 ranged from 0% to 89% , but this criterion was not taken into account for this category). All other associations were classified as negative.
Model-free Approach
For those SNPs that were identified as true positives after applying the BFDP and Venice criteria, we applied the model-free meta-analysis approach described by Thompson et al. (32). This model uses a maximum likelihood estimator, assumes a fixed-effect meta-analysis and is similar to a bivariate meta-analysis of the effects for the heterozygotes and variant homozygotes. It gives an estimate of the lambda (λ), which represents the underlying genetic model (and it represents the heterozygote effect as a proportion of the homozygote effect), with its confidence interval limits. If the confidence interval of λ is too wide, there is not enough power to estimate λ. The value of λ is not restricted. Values equal to 0, 0.5, and 1.0 correspond to the recessive, additive, and dominant genetic models, respectively. Values greater than 1 or smaller than 0 suggest heterosis, which is when the risk of the heterozygotes is higher or lower, respectively, than the risk of either of the homozygote genotypes. However, heterosis is relatively uncommon and λ values greater than 1 or smaller than 0 could represent chance fluctuation, reflecting low statistical power to estimate the magnitude of effect for heterozygotes.
Results
Literature Search and Data Collection
After screening approximately 10 145 titles and abstracts (Supplementary Table 1, available online) dated until June 30, 2010 (or March 31, 2011 for GWAS-identified hits), we identified and extracted data from 635 publications reporting on 445 polymorphisms in 110 different genes. More than 9231 (88%) of these studies were published after 1995, and about half of those were published during the past 3 years.
List of genes and variants that were selected for meta-analysis (sorted by gene pathway)*
| Genes, by category . | Variant . | rs number . | Case vs control subjects (number of samples) . | Ref allele . | Ref allele frequency of case subjects . | Ref allele frequency of control subjects . | Attributable familial risk . | Result of most recent meta-analysis; case subjects/ control subjects (samples) (reference) . | Other meta- analyses . | Source of minor allele frequency . |
|---|---|---|---|---|---|---|---|---|---|---|
| Adhesion molecules | ||||||||||
| CDH1 | C-160A | rs16260 | 7493 vs 7329 (5†) | C | 0.74 | 0.72 | 0.19% | No assoc (63) | — | 1000 Genomes |
| MMP1 | G-1607GG | rs1799750 | 1007 vs 1032 (5) | G | 0.44 | 0.47 | 0.62% | Positive assoc; 1343/1590 (7) (65) | (64) | Current study |
| MMP3 | AAAAA-1612AAAAAA | rs3025058 | 857 vs 932 (4) | 5A | 0.38 | 0.41 | 0.03% | No assoc; 1296/1543 (7) (65) | (64) | Current study |
| MMP9 | 1562C/T ‡ | rs3918242 | 575 vs 836 (4) | C | — | — | — | No assoc; 575/836 (4) (64) | (64) | 1000 Genomes |
| Alcohol metabolism | ||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | His | 0.75 | 0.76 | 0.32% | n/a | — | 1000 Genomes |
| ADH1C | Ile349Val (1045A>G) | rs698 | 3168 vs 6229 (7) | Ile (A) | 0.69 | 0.64 | 0.14% | n/a | — | 1000 Genomes |
| ALDH2 | Glu487Lys | rs671 | 2209 vs 3383 (8) | Glu (G) | 0.76 | 0.74 | 0.02% | Inverse assoc; 1960/3163 (6) (66) | — | 1000 Genomes |
| Angiogenesis | ||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | C | 0.84 | 0.83 | 0.60% | n/a | — | 1000 Genomes |
| VEGF | G634C § | rs2010963 | 1508 vs 1308 (4) | C | — | — | 0.00% || | No assoc; 1508/1308 (4) (131) | — | 1000 Genomes |
| Base-excision repair | ||||||||||
| MGMT | Leu84Phe ¶ | rs12917 | 1524 vs 4646 (5) | C | 0.89 | 0.88 | 0.14% | No assoc; 1524/4646 (5) (137) | — | 1000 Genomes |
| MGMT | Il3143Val ¶ | rs2308321 | 1326 vs 3520 (4) | A | 0.89 | 0.87 | 0.19% | No assoc; 1326/3520 (4) (137) | — | 1000 Genomes |
| MUTYH | G396D | rs36053993 | 26 592 vs 19 207 (15) | G | 0.99 | 0.99 | 0.87% | Positive assoc; 25 616/18 521 (15) (48) | (138,139, (140,141) | Current study |
| MUTYH | Y179C | rs34612342 | 26 370 vs 19 042 (15) | A | 1.00 | 1.00 | 0.00% | Positive assoc; 25 392/18 362 (15) (48) | (138,139, (140,141) | Current study |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | Ser (C) | 0.71 | 0.65 | 0.02% | n/a | — | 1000 Genomes |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11†) | C | 0.91 | 0.88 | 0.16% | No assoc; 1709/3233 (9) (70) | (69) | 1000 Genomes |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | G | 0.94 | 0.95 | 0.04% | No assoc; 931/1547 (4) (70) | — | 1000 Genomes |
| XRCC1 | Arg399Gln | rs25487 | 7247 vs 8786 (12†) | G | 0.67 | 0.67 | 0.19% | No assoc; 2776/4402 (14) (70) | (69) | 1000 Genomes |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10†) | T | 0.72 | 0.71 | 0.00% | No assoc; 3183/3926 (7) (69) | — | 1000 Genomes |
| Inflammation or immune response | ||||||||||
| IL6 | 174G>C | rs1800795 | 6676 vs 7942 (10†,#) | G | 0.61 | 0.60 | 0.05% | n/a | — | 1000 Genomes |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7†) | T | 0.54 | 0.54 | 0.02% | n/a | — | 1000 Genomes |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5†) | A | 0.52 | 0.51 | 0.04% | n/a | — | 1000 Genomes |
| PPAR-ɣ | C1431T | rs3856806 | 5574 vs 7035 (7†) | C | 0.87 | 0.87 | 0.08% | No assoc; 486/941 (3) (106) | (107) | 1000 Genomes |
| PPAR-ɣ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17†,#) | C | 0.88 | 0.88 | 0.06% | Inverse assoc; 6878/9391 (10) (106) | — | 1000 Genomes |
| PTGS2/COX2 | A1195G | rs689466 | 4756 vs 6030 (7†) | A | 0.73 | 0.74 | 0.03% | Positive assoc; 1196/ 1691 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | A1803G | rs4648298 | 4229 vs 4279 (5†) | A | 0.97 | 0.97 | 0.00%** | No assoc; 480/657 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | C427T | rs5275 | 4745 vs 5756 (7†) | T | 0.66 | 0.66 | 0.01% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G306C | rs5277 | 4269 vs 4735 (5†) | G | 0.85 | 0.84 | 0.24% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G765C | rs20417 | 5459 vs 7272 (11†) | G | 0.86 | 0.88 | 0.26% | Positive assoc; 3322/5166 (10) (123) | (142) | 1000 Genomes |
| PTGS2/COX2 | T1532C | rs5273 | 2843 vs 3216 (5†) | val/val | 1.00 | 1.00 | — | No assoc; 670/1113 (4) (142) | — | 1000 Genomes |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9†) | G | 0.80 | 0.81 | 0.02% | No assoc; 1372/1458 (7) (132) | — | Current study |
| NOD2 | 3020incC | rs5743293 | 4222 vs 2988 (8) | G | 0.96 | 0.97 | 0.95% | Positive assoc; 2571/1856 (7) (72) | — | Current study |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6†) | G | 0.99 | 0.99 | — | Positive assoc; 1442/1109 (5) (72) | — | Current study |
| NOD2 | R702W | rs2066844 | 3445 vs 2731 (6†) | C | 0.96 | 0.97 | 0.00% | Positive assoc; 1436/1109 (5) (72) | — | 1000 Genomes |
| Inhibition of cell growth | ||||||||||
| CCND1 | 870A | rs17852153 | 4747 vs 6783 (13) | G | 0.46 | 0.46 | 0.18% | Positive assoc; 2289/3232 (13) (73) | — | Current study |
| TGFB1 | C509T †† | rs1800469 | 994 vs 2335 (5) | T | 0.42 | 0.48 | 1.99% | No assoc; 994/2335 (5) (76) | — | 1000 Genomes |
| TGFBR1 | TGFBR1*6A | rs11466445 | 3217 vs 4539 (8) | C | 0.90 | 0.92 | 0.52% | Positive assoc; 5666/8450 (7) (79) | (77,78) | Current study |
| Insulin related | ||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 19/19 | 0.62 | 0.61 | 0.02% | No assoc; 3672/4125 (4) (117) | (116) | — |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | A | 0.48 | 0.50 | 0.00% | No assoc; 2834/3520 (3) (117) | — | 1000 Genomes |
| Iron metabolism | ||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6†#) | C | 0.92 | 0.93 | 0.25% | n/a | — | 1000 Genomes |
| Lipid metabolism | ||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5†) | e3 | 0.81 | 0.81 | 0.80% | n/a | — | 1000 Genomes |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5†) | e3 | 0.79 | 0.79 | 0.09% | n/a | — | 1000 Genomes |
| Mitotic control | ||||||||||
| STK15 | F31 ‡ | rs2273535 | 4860 vs 4629 (4) | T | 0.75 | 0.76 | 0.31% | Positive assoc; 2302/1769 (3) (143) | — | 1000 Genomes |
| One-carbon metabolism | ||||||||||
| MTHFR | C677T | rs1801133 | 27 372 vs 39 867 (52§) | C | 0.67 | 0.67 | 0.24% | Inverse assoc; 9345/18 887 (37) (56) | (51,52,53, 54,55,88) | 1000 Genomes |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34#) | A | 0.70 | 0.70 | 0.06% | Inverse assoc; 4764/6592 (9) (53) | — | 1000 Genomes |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14#) | A | 0.81 | 0.80 | 0.01% | No assoc; 7804/8184 (9) (110) | (88) | 1000 Genomes |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | A | 0.59 | 0.61 | 0.02% | n/a | — | 1000 Genomes |
| TS | TSER | rs34743033 | 3519 vs 5289 (5) | 3R/3R | 0.57 | 0.57 | 0.24% | n/a | — | Current study |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | ins/ins | 0.67 | 0.67 | 0.03% | n/a | — | Current study |
| Rare, high penetrance | ||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | G | 0.99 | 0.99 | — | No assoc; 3794/4484 (8) (122) | — | Current study |
| APC | D1822V | rs459552 | 6282 vs 7038 (6) | Asp | 0.78 | 0.77 | 0.38% | n/a | — | 1000 Genomes |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7†) | A | 0.71 | 0.71 | 0.02% | n/a | — | 1000 Genomes |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6†) | G | 0.77 | 0.78 | 0.12% | n/a | — | 1000 Genomes |
| Substrate metabolism | ||||||||||
| CYP1A1 | 2454A>G | rs1048943 | 10 274 vs 11 978 (13†,#) | A | 0.91 | 0.92 | 0.18% | Positive assoc; 5336/6226 (13) (87) | (88) | 1000 Genomes |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | T | 0.84 | 0.83 | 0.20% | No assoc; 234/250 (2) (88) | — | 1000 Genomes |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | A | 0.68 | 0.68 | 0.00% | n/a | — | 1000 Genomes |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6†) | C | 0.53 | 0.53 | 0.00% | n/a | — | 1000 Genomes |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6†) | C | 0.86 | 0.86 | 0.95% | n/a | — | HapMap |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6†) | A | 0.93 | 0.94 | 1.85% | n/a | — | 1000 Genomes |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8#) | C | 0.90 | 0.88 | 0.64% | n/a | — | 1000 Genomes |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | G | 0.94 | 0.93 | 1.06% | No assoc; 4979/6012 (10) (133) | — | 1000 Genomes |
| GSTA1 | GSTA1*B allele ‡‡ | — | 1648 vs 2039 (4) | A | — | — | — | No assoc; 1648/2039 (4) (92) | — | — |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | present | 0.48 | 0.49 | — | Positive assoc; 11 998/17 552 (44) (92) | (93,91,89,90,88) | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22†) | IA | 0.71 | 0.72 | 0.09% | No assoc; 5421/7671 (19) (92) | (111,88) | 1000 Genomes |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6†,#) | C | 0.92 | 0.92 | 0.22% | n/a | — | 1000 Genomes |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | present | 0.65 | 0.68 | — | Positive assoc; 8596/13 589 (34) (92) | (88,95,96) | — |
| NAT1 | slow/rapid | n/a | 4791 vs 6628 (15) | slow | 0.69 | 0.68 | 0.12% | No assoc; 520/433 (3) (88) | — | — |
| NAT2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | slow | 0.67 | 0.66 | 0.04% | No assoc; 6741/8015 (18) (88) | (108,51,109) | — |
| NQO1 | Pro187Ser (C609T) | rs1800566 | 5084 vs 5932 (8) | C | 0.81 | 0.79 | 0.00% | Positive assoc; 1783/2494 (6) (130) | — | 1000 Genomes |
| Tumor suppressor genes | ||||||||||
| TP53 | Arg72Pro §§ | rs1042522 | 7414 vs 9872 (27) | G | — | — | 0.01% | No assoc; 7414/9872 (27) (119) | (118,120,121) | 1000 Genomes |
| TP53 | intron 3 16bp |||| | rs17878362 | 1637 vs 1874 (5) | Del | — | — | 0.00% | No assoc; 1637/1874 (5) (144) | — | — |
| MDM2 | 309 T/G ¶¶ | rs2279744 | 2543 vs 2115 (7) | G | 0.46 | 0.43 | 0.42% | No assoc; 2543/2115 (7) (145) | (146) | 1000 Genomes |
| Vitamin D and calcium metabolism | ||||||||||
| VDR | BsmI (60890GA) | rs1544410 | 5607 vs 6202 (7) | G | 0.63 | 0.60 | 0.12%## | Inverse assoc; 3285/1497 (4) (147) | — | 1000 Genomes |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9#) | C | 0.61 | 0.60 | 0.00% | No assoc; 1331/2943 (5) (147) | — | 1000 Genomes |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | T | 0.68 | 0.70 | 0.01% | n/a | — | 1000 Genomes |
| Common low penetrance | ||||||||||
| SMAD7 | rs4939827 | rs4939827 | 37 650 vs 36 154 (13#) | T | 0.55 | 0.51 | 0.64% | n/a | — | 1000 Genomes |
| SMAD7 | rs12953717 | rs12953717 | 33 771 vs 32 364 (11#) | C | 0.63 | 0.65 | 0.30% | n/a | — | 1000 Genomes |
| SMAD7 | rs4464148 | rs4464148 | 15 999 vs 15 216 (7†) | T | 0.62 | 0.66 | 0.47% | n/a | — | 1000 Genomes |
| 8q24 | rs6983267 | rs6983267 | 40 604 vs 42 672 (19) | A | 0.48 | 0.51 | 1.06% | Positive assoc; (17) (148) | — | 1000 Genomes |
| 8q24 | rs10505477 | rs10505477 | 18 580 vs 20 147 (14) | C | 0.46 | 0.49 | 0.59% | n/a | — | 1000 Genomes |
| 9p24 | rs719725 | rs719725 | 13 290 vs 14 774 (13) | C | 0.37 | 0.39 | 0.16% | Positive assoc; 14 064/15 933 (17) (102) | — | 1000 Genomes |
| 19q13.1 | rs10411210 | rs10411210 | 25 607 vs 26 477 (17) | C | 0.89 | 0.88 | 0.09% | n/a | — | 1000 Genomes |
| 16q22.1 | rs9929218 | rs9929218 | 26 191 vs 27 409 (18) | G | 0.74 | 0.72 | 0.23% | n/a | — | 1000 Genomes |
| 15q14 | rs4779584 | rs4779584 | 13 656 vs 12 635 (9) | C | 0.64 | 0.65 | 0.84% | n/a | — | 1000 Genomes |
| 1q41 | rs6691170 | rs6691170 | 17 740 vs 19 776 (11) | G | 0.62 | 0.64 | 0.18% | n/a | — | 1000 Genomes |
| 3q26.2 | rs10936599 | rs10936599 | 17 802 vs 19 795 (11) | C | 0.77 | 0.75 | 0.12% | n/a | — | 1000 Genomes |
| 12q13.13 | rs11169552 | rs11169552 | 17 148 vs 19 739 (11) | C | 0.74 | 0.72 | 0.76% | n/a | — | 1000 Genomes |
| 20q13.33 | rs4925386 | rs4925386 | 17 847 vs 19 832 (11) | C | 0.71 | 0.68 | 0.39% | n/a | — | 1000 Genomes |
| 14q22.2 | rs4444235 | rs4444235 | 18 607 vs 19 576 (13) | T | 0.53 | 0.55 | 0.21% | n/a | — | 1000 Genomes |
| 20p12.3 | rs961253 | rs961253 | 18 118 vs 19 006 (13) | C | 0.66 | 0.68 | 0.22% | n/a | — | 1000 Genomes |
| 8q23.3 | rs16892766 | rs16892766 | 17 180 vs 17 840 (4†) | A | 0.88 | 0.90 | 0.10% | n/a | — | 1000 Genomes |
| 10p14 | rs10795668 | rs10795668 | 20 026 vs 20 682 (6†) | G | 0.72 | 0.69 | 0.52% | n/a | — | 1000 Genomes |
| 11q23.1 | rs3802842 | rs3802842 | 33 004 vs 31 654 (14) | A | 0.67 | 0.70 | 0.37% | n/a | — | 1000 Genomes |
| Genes, by category . | Variant . | rs number . | Case vs control subjects (number of samples) . | Ref allele . | Ref allele frequency of case subjects . | Ref allele frequency of control subjects . | Attributable familial risk . | Result of most recent meta-analysis; case subjects/ control subjects (samples) (reference) . | Other meta- analyses . | Source of minor allele frequency . |
|---|---|---|---|---|---|---|---|---|---|---|
| Adhesion molecules | ||||||||||
| CDH1 | C-160A | rs16260 | 7493 vs 7329 (5†) | C | 0.74 | 0.72 | 0.19% | No assoc (63) | — | 1000 Genomes |
| MMP1 | G-1607GG | rs1799750 | 1007 vs 1032 (5) | G | 0.44 | 0.47 | 0.62% | Positive assoc; 1343/1590 (7) (65) | (64) | Current study |
| MMP3 | AAAAA-1612AAAAAA | rs3025058 | 857 vs 932 (4) | 5A | 0.38 | 0.41 | 0.03% | No assoc; 1296/1543 (7) (65) | (64) | Current study |
| MMP9 | 1562C/T ‡ | rs3918242 | 575 vs 836 (4) | C | — | — | — | No assoc; 575/836 (4) (64) | (64) | 1000 Genomes |
| Alcohol metabolism | ||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | His | 0.75 | 0.76 | 0.32% | n/a | — | 1000 Genomes |
| ADH1C | Ile349Val (1045A>G) | rs698 | 3168 vs 6229 (7) | Ile (A) | 0.69 | 0.64 | 0.14% | n/a | — | 1000 Genomes |
| ALDH2 | Glu487Lys | rs671 | 2209 vs 3383 (8) | Glu (G) | 0.76 | 0.74 | 0.02% | Inverse assoc; 1960/3163 (6) (66) | — | 1000 Genomes |
| Angiogenesis | ||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | C | 0.84 | 0.83 | 0.60% | n/a | — | 1000 Genomes |
| VEGF | G634C § | rs2010963 | 1508 vs 1308 (4) | C | — | — | 0.00% || | No assoc; 1508/1308 (4) (131) | — | 1000 Genomes |
| Base-excision repair | ||||||||||
| MGMT | Leu84Phe ¶ | rs12917 | 1524 vs 4646 (5) | C | 0.89 | 0.88 | 0.14% | No assoc; 1524/4646 (5) (137) | — | 1000 Genomes |
| MGMT | Il3143Val ¶ | rs2308321 | 1326 vs 3520 (4) | A | 0.89 | 0.87 | 0.19% | No assoc; 1326/3520 (4) (137) | — | 1000 Genomes |
| MUTYH | G396D | rs36053993 | 26 592 vs 19 207 (15) | G | 0.99 | 0.99 | 0.87% | Positive assoc; 25 616/18 521 (15) (48) | (138,139, (140,141) | Current study |
| MUTYH | Y179C | rs34612342 | 26 370 vs 19 042 (15) | A | 1.00 | 1.00 | 0.00% | Positive assoc; 25 392/18 362 (15) (48) | (138,139, (140,141) | Current study |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | Ser (C) | 0.71 | 0.65 | 0.02% | n/a | — | 1000 Genomes |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11†) | C | 0.91 | 0.88 | 0.16% | No assoc; 1709/3233 (9) (70) | (69) | 1000 Genomes |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | G | 0.94 | 0.95 | 0.04% | No assoc; 931/1547 (4) (70) | — | 1000 Genomes |
| XRCC1 | Arg399Gln | rs25487 | 7247 vs 8786 (12†) | G | 0.67 | 0.67 | 0.19% | No assoc; 2776/4402 (14) (70) | (69) | 1000 Genomes |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10†) | T | 0.72 | 0.71 | 0.00% | No assoc; 3183/3926 (7) (69) | — | 1000 Genomes |
| Inflammation or immune response | ||||||||||
| IL6 | 174G>C | rs1800795 | 6676 vs 7942 (10†,#) | G | 0.61 | 0.60 | 0.05% | n/a | — | 1000 Genomes |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7†) | T | 0.54 | 0.54 | 0.02% | n/a | — | 1000 Genomes |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5†) | A | 0.52 | 0.51 | 0.04% | n/a | — | 1000 Genomes |
| PPAR-ɣ | C1431T | rs3856806 | 5574 vs 7035 (7†) | C | 0.87 | 0.87 | 0.08% | No assoc; 486/941 (3) (106) | (107) | 1000 Genomes |
| PPAR-ɣ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17†,#) | C | 0.88 | 0.88 | 0.06% | Inverse assoc; 6878/9391 (10) (106) | — | 1000 Genomes |
| PTGS2/COX2 | A1195G | rs689466 | 4756 vs 6030 (7†) | A | 0.73 | 0.74 | 0.03% | Positive assoc; 1196/ 1691 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | A1803G | rs4648298 | 4229 vs 4279 (5†) | A | 0.97 | 0.97 | 0.00%** | No assoc; 480/657 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | C427T | rs5275 | 4745 vs 5756 (7†) | T | 0.66 | 0.66 | 0.01% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G306C | rs5277 | 4269 vs 4735 (5†) | G | 0.85 | 0.84 | 0.24% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G765C | rs20417 | 5459 vs 7272 (11†) | G | 0.86 | 0.88 | 0.26% | Positive assoc; 3322/5166 (10) (123) | (142) | 1000 Genomes |
| PTGS2/COX2 | T1532C | rs5273 | 2843 vs 3216 (5†) | val/val | 1.00 | 1.00 | — | No assoc; 670/1113 (4) (142) | — | 1000 Genomes |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9†) | G | 0.80 | 0.81 | 0.02% | No assoc; 1372/1458 (7) (132) | — | Current study |
| NOD2 | 3020incC | rs5743293 | 4222 vs 2988 (8) | G | 0.96 | 0.97 | 0.95% | Positive assoc; 2571/1856 (7) (72) | — | Current study |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6†) | G | 0.99 | 0.99 | — | Positive assoc; 1442/1109 (5) (72) | — | Current study |
| NOD2 | R702W | rs2066844 | 3445 vs 2731 (6†) | C | 0.96 | 0.97 | 0.00% | Positive assoc; 1436/1109 (5) (72) | — | 1000 Genomes |
| Inhibition of cell growth | ||||||||||
| CCND1 | 870A | rs17852153 | 4747 vs 6783 (13) | G | 0.46 | 0.46 | 0.18% | Positive assoc; 2289/3232 (13) (73) | — | Current study |
| TGFB1 | C509T †† | rs1800469 | 994 vs 2335 (5) | T | 0.42 | 0.48 | 1.99% | No assoc; 994/2335 (5) (76) | — | 1000 Genomes |
| TGFBR1 | TGFBR1*6A | rs11466445 | 3217 vs 4539 (8) | C | 0.90 | 0.92 | 0.52% | Positive assoc; 5666/8450 (7) (79) | (77,78) | Current study |
| Insulin related | ||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 19/19 | 0.62 | 0.61 | 0.02% | No assoc; 3672/4125 (4) (117) | (116) | — |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | A | 0.48 | 0.50 | 0.00% | No assoc; 2834/3520 (3) (117) | — | 1000 Genomes |
| Iron metabolism | ||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6†#) | C | 0.92 | 0.93 | 0.25% | n/a | — | 1000 Genomes |
| Lipid metabolism | ||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5†) | e3 | 0.81 | 0.81 | 0.80% | n/a | — | 1000 Genomes |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5†) | e3 | 0.79 | 0.79 | 0.09% | n/a | — | 1000 Genomes |
| Mitotic control | ||||||||||
| STK15 | F31 ‡ | rs2273535 | 4860 vs 4629 (4) | T | 0.75 | 0.76 | 0.31% | Positive assoc; 2302/1769 (3) (143) | — | 1000 Genomes |
| One-carbon metabolism | ||||||||||
| MTHFR | C677T | rs1801133 | 27 372 vs 39 867 (52§) | C | 0.67 | 0.67 | 0.24% | Inverse assoc; 9345/18 887 (37) (56) | (51,52,53, 54,55,88) | 1000 Genomes |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34#) | A | 0.70 | 0.70 | 0.06% | Inverse assoc; 4764/6592 (9) (53) | — | 1000 Genomes |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14#) | A | 0.81 | 0.80 | 0.01% | No assoc; 7804/8184 (9) (110) | (88) | 1000 Genomes |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | A | 0.59 | 0.61 | 0.02% | n/a | — | 1000 Genomes |
| TS | TSER | rs34743033 | 3519 vs 5289 (5) | 3R/3R | 0.57 | 0.57 | 0.24% | n/a | — | Current study |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | ins/ins | 0.67 | 0.67 | 0.03% | n/a | — | Current study |
| Rare, high penetrance | ||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | G | 0.99 | 0.99 | — | No assoc; 3794/4484 (8) (122) | — | Current study |
| APC | D1822V | rs459552 | 6282 vs 7038 (6) | Asp | 0.78 | 0.77 | 0.38% | n/a | — | 1000 Genomes |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7†) | A | 0.71 | 0.71 | 0.02% | n/a | — | 1000 Genomes |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6†) | G | 0.77 | 0.78 | 0.12% | n/a | — | 1000 Genomes |
| Substrate metabolism | ||||||||||
| CYP1A1 | 2454A>G | rs1048943 | 10 274 vs 11 978 (13†,#) | A | 0.91 | 0.92 | 0.18% | Positive assoc; 5336/6226 (13) (87) | (88) | 1000 Genomes |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | T | 0.84 | 0.83 | 0.20% | No assoc; 234/250 (2) (88) | — | 1000 Genomes |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | A | 0.68 | 0.68 | 0.00% | n/a | — | 1000 Genomes |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6†) | C | 0.53 | 0.53 | 0.00% | n/a | — | 1000 Genomes |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6†) | C | 0.86 | 0.86 | 0.95% | n/a | — | HapMap |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6†) | A | 0.93 | 0.94 | 1.85% | n/a | — | 1000 Genomes |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8#) | C | 0.90 | 0.88 | 0.64% | n/a | — | 1000 Genomes |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | G | 0.94 | 0.93 | 1.06% | No assoc; 4979/6012 (10) (133) | — | 1000 Genomes |
| GSTA1 | GSTA1*B allele ‡‡ | — | 1648 vs 2039 (4) | A | — | — | — | No assoc; 1648/2039 (4) (92) | — | — |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | present | 0.48 | 0.49 | — | Positive assoc; 11 998/17 552 (44) (92) | (93,91,89,90,88) | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22†) | IA | 0.71 | 0.72 | 0.09% | No assoc; 5421/7671 (19) (92) | (111,88) | 1000 Genomes |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6†,#) | C | 0.92 | 0.92 | 0.22% | n/a | — | 1000 Genomes |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | present | 0.65 | 0.68 | — | Positive assoc; 8596/13 589 (34) (92) | (88,95,96) | — |
| NAT1 | slow/rapid | n/a | 4791 vs 6628 (15) | slow | 0.69 | 0.68 | 0.12% | No assoc; 520/433 (3) (88) | — | — |
| NAT2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | slow | 0.67 | 0.66 | 0.04% | No assoc; 6741/8015 (18) (88) | (108,51,109) | — |
| NQO1 | Pro187Ser (C609T) | rs1800566 | 5084 vs 5932 (8) | C | 0.81 | 0.79 | 0.00% | Positive assoc; 1783/2494 (6) (130) | — | 1000 Genomes |
| Tumor suppressor genes | ||||||||||
| TP53 | Arg72Pro §§ | rs1042522 | 7414 vs 9872 (27) | G | — | — | 0.01% | No assoc; 7414/9872 (27) (119) | (118,120,121) | 1000 Genomes |
| TP53 | intron 3 16bp |||| | rs17878362 | 1637 vs 1874 (5) | Del | — | — | 0.00% | No assoc; 1637/1874 (5) (144) | — | — |
| MDM2 | 309 T/G ¶¶ | rs2279744 | 2543 vs 2115 (7) | G | 0.46 | 0.43 | 0.42% | No assoc; 2543/2115 (7) (145) | (146) | 1000 Genomes |
| Vitamin D and calcium metabolism | ||||||||||
| VDR | BsmI (60890GA) | rs1544410 | 5607 vs 6202 (7) | G | 0.63 | 0.60 | 0.12%## | Inverse assoc; 3285/1497 (4) (147) | — | 1000 Genomes |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9#) | C | 0.61 | 0.60 | 0.00% | No assoc; 1331/2943 (5) (147) | — | 1000 Genomes |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | T | 0.68 | 0.70 | 0.01% | n/a | — | 1000 Genomes |
| Common low penetrance | ||||||||||
| SMAD7 | rs4939827 | rs4939827 | 37 650 vs 36 154 (13#) | T | 0.55 | 0.51 | 0.64% | n/a | — | 1000 Genomes |
| SMAD7 | rs12953717 | rs12953717 | 33 771 vs 32 364 (11#) | C | 0.63 | 0.65 | 0.30% | n/a | — | 1000 Genomes |
| SMAD7 | rs4464148 | rs4464148 | 15 999 vs 15 216 (7†) | T | 0.62 | 0.66 | 0.47% | n/a | — | 1000 Genomes |
| 8q24 | rs6983267 | rs6983267 | 40 604 vs 42 672 (19) | A | 0.48 | 0.51 | 1.06% | Positive assoc; (17) (148) | — | 1000 Genomes |
| 8q24 | rs10505477 | rs10505477 | 18 580 vs 20 147 (14) | C | 0.46 | 0.49 | 0.59% | n/a | — | 1000 Genomes |
| 9p24 | rs719725 | rs719725 | 13 290 vs 14 774 (13) | C | 0.37 | 0.39 | 0.16% | Positive assoc; 14 064/15 933 (17) (102) | — | 1000 Genomes |
| 19q13.1 | rs10411210 | rs10411210 | 25 607 vs 26 477 (17) | C | 0.89 | 0.88 | 0.09% | n/a | — | 1000 Genomes |
| 16q22.1 | rs9929218 | rs9929218 | 26 191 vs 27 409 (18) | G | 0.74 | 0.72 | 0.23% | n/a | — | 1000 Genomes |
| 15q14 | rs4779584 | rs4779584 | 13 656 vs 12 635 (9) | C | 0.64 | 0.65 | 0.84% | n/a | — | 1000 Genomes |
| 1q41 | rs6691170 | rs6691170 | 17 740 vs 19 776 (11) | G | 0.62 | 0.64 | 0.18% | n/a | — | 1000 Genomes |
| 3q26.2 | rs10936599 | rs10936599 | 17 802 vs 19 795 (11) | C | 0.77 | 0.75 | 0.12% | n/a | — | 1000 Genomes |
| 12q13.13 | rs11169552 | rs11169552 | 17 148 vs 19 739 (11) | C | 0.74 | 0.72 | 0.76% | n/a | — | 1000 Genomes |
| 20q13.33 | rs4925386 | rs4925386 | 17 847 vs 19 832 (11) | C | 0.71 | 0.68 | 0.39% | n/a | — | 1000 Genomes |
| 14q22.2 | rs4444235 | rs4444235 | 18 607 vs 19 576 (13) | T | 0.53 | 0.55 | 0.21% | n/a | — | 1000 Genomes |
| 20p12.3 | rs961253 | rs961253 | 18 118 vs 19 006 (13) | C | 0.66 | 0.68 | 0.22% | n/a | — | 1000 Genomes |
| 8q23.3 | rs16892766 | rs16892766 | 17 180 vs 17 840 (4†) | A | 0.88 | 0.90 | 0.10% | n/a | — | 1000 Genomes |
| 10p14 | rs10795668 | rs10795668 | 20 026 vs 20 682 (6†) | G | 0.72 | 0.69 | 0.52% | n/a | — | 1000 Genomes |
| 11q23.1 | rs3802842 | rs3802842 | 33 004 vs 31 654 (14) | A | 0.67 | 0.70 | 0.37% | n/a | — | 1000 Genomes |
* n/a = not applicable. See Supplementary Table 1 (available online) for gene names.
† Includes unpublished data from SOCCS.
‡ McColgan and Sharma 2009 (64).
§ Liu et al. 2011 (131).
|| ref allele frequency taken from 1000 Genomes data.
¶ Zhong et al. 2010 (137).
# Includes unpublished data from Ontario.
** OR for homozygote estimated as square of OR for heterozygotes.
†† Fang et al. 2010 (76).
‡‡ Economopoulos and Sergentanis 2010 (92).
§§ Economopoulos et al. 2010 (119).
|||| Hu et al. 2010 (144).
¶¶ Tomlinson 2008 was based on 10 samples.
## Based on the white-only analysis.
List of genes and variants that were selected for meta-analysis (sorted by gene pathway)*
| Genes, by category . | Variant . | rs number . | Case vs control subjects (number of samples) . | Ref allele . | Ref allele frequency of case subjects . | Ref allele frequency of control subjects . | Attributable familial risk . | Result of most recent meta-analysis; case subjects/ control subjects (samples) (reference) . | Other meta- analyses . | Source of minor allele frequency . |
|---|---|---|---|---|---|---|---|---|---|---|
| Adhesion molecules | ||||||||||
| CDH1 | C-160A | rs16260 | 7493 vs 7329 (5†) | C | 0.74 | 0.72 | 0.19% | No assoc (63) | — | 1000 Genomes |
| MMP1 | G-1607GG | rs1799750 | 1007 vs 1032 (5) | G | 0.44 | 0.47 | 0.62% | Positive assoc; 1343/1590 (7) (65) | (64) | Current study |
| MMP3 | AAAAA-1612AAAAAA | rs3025058 | 857 vs 932 (4) | 5A | 0.38 | 0.41 | 0.03% | No assoc; 1296/1543 (7) (65) | (64) | Current study |
| MMP9 | 1562C/T ‡ | rs3918242 | 575 vs 836 (4) | C | — | — | — | No assoc; 575/836 (4) (64) | (64) | 1000 Genomes |
| Alcohol metabolism | ||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | His | 0.75 | 0.76 | 0.32% | n/a | — | 1000 Genomes |
| ADH1C | Ile349Val (1045A>G) | rs698 | 3168 vs 6229 (7) | Ile (A) | 0.69 | 0.64 | 0.14% | n/a | — | 1000 Genomes |
| ALDH2 | Glu487Lys | rs671 | 2209 vs 3383 (8) | Glu (G) | 0.76 | 0.74 | 0.02% | Inverse assoc; 1960/3163 (6) (66) | — | 1000 Genomes |
| Angiogenesis | ||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | C | 0.84 | 0.83 | 0.60% | n/a | — | 1000 Genomes |
| VEGF | G634C § | rs2010963 | 1508 vs 1308 (4) | C | — | — | 0.00% || | No assoc; 1508/1308 (4) (131) | — | 1000 Genomes |
| Base-excision repair | ||||||||||
| MGMT | Leu84Phe ¶ | rs12917 | 1524 vs 4646 (5) | C | 0.89 | 0.88 | 0.14% | No assoc; 1524/4646 (5) (137) | — | 1000 Genomes |
| MGMT | Il3143Val ¶ | rs2308321 | 1326 vs 3520 (4) | A | 0.89 | 0.87 | 0.19% | No assoc; 1326/3520 (4) (137) | — | 1000 Genomes |
| MUTYH | G396D | rs36053993 | 26 592 vs 19 207 (15) | G | 0.99 | 0.99 | 0.87% | Positive assoc; 25 616/18 521 (15) (48) | (138,139, (140,141) | Current study |
| MUTYH | Y179C | rs34612342 | 26 370 vs 19 042 (15) | A | 1.00 | 1.00 | 0.00% | Positive assoc; 25 392/18 362 (15) (48) | (138,139, (140,141) | Current study |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | Ser (C) | 0.71 | 0.65 | 0.02% | n/a | — | 1000 Genomes |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11†) | C | 0.91 | 0.88 | 0.16% | No assoc; 1709/3233 (9) (70) | (69) | 1000 Genomes |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | G | 0.94 | 0.95 | 0.04% | No assoc; 931/1547 (4) (70) | — | 1000 Genomes |
| XRCC1 | Arg399Gln | rs25487 | 7247 vs 8786 (12†) | G | 0.67 | 0.67 | 0.19% | No assoc; 2776/4402 (14) (70) | (69) | 1000 Genomes |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10†) | T | 0.72 | 0.71 | 0.00% | No assoc; 3183/3926 (7) (69) | — | 1000 Genomes |
| Inflammation or immune response | ||||||||||
| IL6 | 174G>C | rs1800795 | 6676 vs 7942 (10†,#) | G | 0.61 | 0.60 | 0.05% | n/a | — | 1000 Genomes |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7†) | T | 0.54 | 0.54 | 0.02% | n/a | — | 1000 Genomes |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5†) | A | 0.52 | 0.51 | 0.04% | n/a | — | 1000 Genomes |
| PPAR-ɣ | C1431T | rs3856806 | 5574 vs 7035 (7†) | C | 0.87 | 0.87 | 0.08% | No assoc; 486/941 (3) (106) | (107) | 1000 Genomes |
| PPAR-ɣ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17†,#) | C | 0.88 | 0.88 | 0.06% | Inverse assoc; 6878/9391 (10) (106) | — | 1000 Genomes |
| PTGS2/COX2 | A1195G | rs689466 | 4756 vs 6030 (7†) | A | 0.73 | 0.74 | 0.03% | Positive assoc; 1196/ 1691 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | A1803G | rs4648298 | 4229 vs 4279 (5†) | A | 0.97 | 0.97 | 0.00%** | No assoc; 480/657 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | C427T | rs5275 | 4745 vs 5756 (7†) | T | 0.66 | 0.66 | 0.01% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G306C | rs5277 | 4269 vs 4735 (5†) | G | 0.85 | 0.84 | 0.24% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G765C | rs20417 | 5459 vs 7272 (11†) | G | 0.86 | 0.88 | 0.26% | Positive assoc; 3322/5166 (10) (123) | (142) | 1000 Genomes |
| PTGS2/COX2 | T1532C | rs5273 | 2843 vs 3216 (5†) | val/val | 1.00 | 1.00 | — | No assoc; 670/1113 (4) (142) | — | 1000 Genomes |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9†) | G | 0.80 | 0.81 | 0.02% | No assoc; 1372/1458 (7) (132) | — | Current study |
| NOD2 | 3020incC | rs5743293 | 4222 vs 2988 (8) | G | 0.96 | 0.97 | 0.95% | Positive assoc; 2571/1856 (7) (72) | — | Current study |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6†) | G | 0.99 | 0.99 | — | Positive assoc; 1442/1109 (5) (72) | — | Current study |
| NOD2 | R702W | rs2066844 | 3445 vs 2731 (6†) | C | 0.96 | 0.97 | 0.00% | Positive assoc; 1436/1109 (5) (72) | — | 1000 Genomes |
| Inhibition of cell growth | ||||||||||
| CCND1 | 870A | rs17852153 | 4747 vs 6783 (13) | G | 0.46 | 0.46 | 0.18% | Positive assoc; 2289/3232 (13) (73) | — | Current study |
| TGFB1 | C509T †† | rs1800469 | 994 vs 2335 (5) | T | 0.42 | 0.48 | 1.99% | No assoc; 994/2335 (5) (76) | — | 1000 Genomes |
| TGFBR1 | TGFBR1*6A | rs11466445 | 3217 vs 4539 (8) | C | 0.90 | 0.92 | 0.52% | Positive assoc; 5666/8450 (7) (79) | (77,78) | Current study |
| Insulin related | ||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 19/19 | 0.62 | 0.61 | 0.02% | No assoc; 3672/4125 (4) (117) | (116) | — |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | A | 0.48 | 0.50 | 0.00% | No assoc; 2834/3520 (3) (117) | — | 1000 Genomes |
| Iron metabolism | ||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6†#) | C | 0.92 | 0.93 | 0.25% | n/a | — | 1000 Genomes |
| Lipid metabolism | ||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5†) | e3 | 0.81 | 0.81 | 0.80% | n/a | — | 1000 Genomes |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5†) | e3 | 0.79 | 0.79 | 0.09% | n/a | — | 1000 Genomes |
| Mitotic control | ||||||||||
| STK15 | F31 ‡ | rs2273535 | 4860 vs 4629 (4) | T | 0.75 | 0.76 | 0.31% | Positive assoc; 2302/1769 (3) (143) | — | 1000 Genomes |
| One-carbon metabolism | ||||||||||
| MTHFR | C677T | rs1801133 | 27 372 vs 39 867 (52§) | C | 0.67 | 0.67 | 0.24% | Inverse assoc; 9345/18 887 (37) (56) | (51,52,53, 54,55,88) | 1000 Genomes |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34#) | A | 0.70 | 0.70 | 0.06% | Inverse assoc; 4764/6592 (9) (53) | — | 1000 Genomes |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14#) | A | 0.81 | 0.80 | 0.01% | No assoc; 7804/8184 (9) (110) | (88) | 1000 Genomes |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | A | 0.59 | 0.61 | 0.02% | n/a | — | 1000 Genomes |
| TS | TSER | rs34743033 | 3519 vs 5289 (5) | 3R/3R | 0.57 | 0.57 | 0.24% | n/a | — | Current study |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | ins/ins | 0.67 | 0.67 | 0.03% | n/a | — | Current study |
| Rare, high penetrance | ||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | G | 0.99 | 0.99 | — | No assoc; 3794/4484 (8) (122) | — | Current study |
| APC | D1822V | rs459552 | 6282 vs 7038 (6) | Asp | 0.78 | 0.77 | 0.38% | n/a | — | 1000 Genomes |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7†) | A | 0.71 | 0.71 | 0.02% | n/a | — | 1000 Genomes |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6†) | G | 0.77 | 0.78 | 0.12% | n/a | — | 1000 Genomes |
| Substrate metabolism | ||||||||||
| CYP1A1 | 2454A>G | rs1048943 | 10 274 vs 11 978 (13†,#) | A | 0.91 | 0.92 | 0.18% | Positive assoc; 5336/6226 (13) (87) | (88) | 1000 Genomes |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | T | 0.84 | 0.83 | 0.20% | No assoc; 234/250 (2) (88) | — | 1000 Genomes |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | A | 0.68 | 0.68 | 0.00% | n/a | — | 1000 Genomes |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6†) | C | 0.53 | 0.53 | 0.00% | n/a | — | 1000 Genomes |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6†) | C | 0.86 | 0.86 | 0.95% | n/a | — | HapMap |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6†) | A | 0.93 | 0.94 | 1.85% | n/a | — | 1000 Genomes |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8#) | C | 0.90 | 0.88 | 0.64% | n/a | — | 1000 Genomes |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | G | 0.94 | 0.93 | 1.06% | No assoc; 4979/6012 (10) (133) | — | 1000 Genomes |
| GSTA1 | GSTA1*B allele ‡‡ | — | 1648 vs 2039 (4) | A | — | — | — | No assoc; 1648/2039 (4) (92) | — | — |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | present | 0.48 | 0.49 | — | Positive assoc; 11 998/17 552 (44) (92) | (93,91,89,90,88) | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22†) | IA | 0.71 | 0.72 | 0.09% | No assoc; 5421/7671 (19) (92) | (111,88) | 1000 Genomes |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6†,#) | C | 0.92 | 0.92 | 0.22% | n/a | — | 1000 Genomes |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | present | 0.65 | 0.68 | — | Positive assoc; 8596/13 589 (34) (92) | (88,95,96) | — |
| NAT1 | slow/rapid | n/a | 4791 vs 6628 (15) | slow | 0.69 | 0.68 | 0.12% | No assoc; 520/433 (3) (88) | — | — |
| NAT2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | slow | 0.67 | 0.66 | 0.04% | No assoc; 6741/8015 (18) (88) | (108,51,109) | — |
| NQO1 | Pro187Ser (C609T) | rs1800566 | 5084 vs 5932 (8) | C | 0.81 | 0.79 | 0.00% | Positive assoc; 1783/2494 (6) (130) | — | 1000 Genomes |
| Tumor suppressor genes | ||||||||||
| TP53 | Arg72Pro §§ | rs1042522 | 7414 vs 9872 (27) | G | — | — | 0.01% | No assoc; 7414/9872 (27) (119) | (118,120,121) | 1000 Genomes |
| TP53 | intron 3 16bp |||| | rs17878362 | 1637 vs 1874 (5) | Del | — | — | 0.00% | No assoc; 1637/1874 (5) (144) | — | — |
| MDM2 | 309 T/G ¶¶ | rs2279744 | 2543 vs 2115 (7) | G | 0.46 | 0.43 | 0.42% | No assoc; 2543/2115 (7) (145) | (146) | 1000 Genomes |
| Vitamin D and calcium metabolism | ||||||||||
| VDR | BsmI (60890GA) | rs1544410 | 5607 vs 6202 (7) | G | 0.63 | 0.60 | 0.12%## | Inverse assoc; 3285/1497 (4) (147) | — | 1000 Genomes |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9#) | C | 0.61 | 0.60 | 0.00% | No assoc; 1331/2943 (5) (147) | — | 1000 Genomes |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | T | 0.68 | 0.70 | 0.01% | n/a | — | 1000 Genomes |
| Common low penetrance | ||||||||||
| SMAD7 | rs4939827 | rs4939827 | 37 650 vs 36 154 (13#) | T | 0.55 | 0.51 | 0.64% | n/a | — | 1000 Genomes |
| SMAD7 | rs12953717 | rs12953717 | 33 771 vs 32 364 (11#) | C | 0.63 | 0.65 | 0.30% | n/a | — | 1000 Genomes |
| SMAD7 | rs4464148 | rs4464148 | 15 999 vs 15 216 (7†) | T | 0.62 | 0.66 | 0.47% | n/a | — | 1000 Genomes |
| 8q24 | rs6983267 | rs6983267 | 40 604 vs 42 672 (19) | A | 0.48 | 0.51 | 1.06% | Positive assoc; (17) (148) | — | 1000 Genomes |
| 8q24 | rs10505477 | rs10505477 | 18 580 vs 20 147 (14) | C | 0.46 | 0.49 | 0.59% | n/a | — | 1000 Genomes |
| 9p24 | rs719725 | rs719725 | 13 290 vs 14 774 (13) | C | 0.37 | 0.39 | 0.16% | Positive assoc; 14 064/15 933 (17) (102) | — | 1000 Genomes |
| 19q13.1 | rs10411210 | rs10411210 | 25 607 vs 26 477 (17) | C | 0.89 | 0.88 | 0.09% | n/a | — | 1000 Genomes |
| 16q22.1 | rs9929218 | rs9929218 | 26 191 vs 27 409 (18) | G | 0.74 | 0.72 | 0.23% | n/a | — | 1000 Genomes |
| 15q14 | rs4779584 | rs4779584 | 13 656 vs 12 635 (9) | C | 0.64 | 0.65 | 0.84% | n/a | — | 1000 Genomes |
| 1q41 | rs6691170 | rs6691170 | 17 740 vs 19 776 (11) | G | 0.62 | 0.64 | 0.18% | n/a | — | 1000 Genomes |
| 3q26.2 | rs10936599 | rs10936599 | 17 802 vs 19 795 (11) | C | 0.77 | 0.75 | 0.12% | n/a | — | 1000 Genomes |
| 12q13.13 | rs11169552 | rs11169552 | 17 148 vs 19 739 (11) | C | 0.74 | 0.72 | 0.76% | n/a | — | 1000 Genomes |
| 20q13.33 | rs4925386 | rs4925386 | 17 847 vs 19 832 (11) | C | 0.71 | 0.68 | 0.39% | n/a | — | 1000 Genomes |
| 14q22.2 | rs4444235 | rs4444235 | 18 607 vs 19 576 (13) | T | 0.53 | 0.55 | 0.21% | n/a | — | 1000 Genomes |
| 20p12.3 | rs961253 | rs961253 | 18 118 vs 19 006 (13) | C | 0.66 | 0.68 | 0.22% | n/a | — | 1000 Genomes |
| 8q23.3 | rs16892766 | rs16892766 | 17 180 vs 17 840 (4†) | A | 0.88 | 0.90 | 0.10% | n/a | — | 1000 Genomes |
| 10p14 | rs10795668 | rs10795668 | 20 026 vs 20 682 (6†) | G | 0.72 | 0.69 | 0.52% | n/a | — | 1000 Genomes |
| 11q23.1 | rs3802842 | rs3802842 | 33 004 vs 31 654 (14) | A | 0.67 | 0.70 | 0.37% | n/a | — | 1000 Genomes |
| Genes, by category . | Variant . | rs number . | Case vs control subjects (number of samples) . | Ref allele . | Ref allele frequency of case subjects . | Ref allele frequency of control subjects . | Attributable familial risk . | Result of most recent meta-analysis; case subjects/ control subjects (samples) (reference) . | Other meta- analyses . | Source of minor allele frequency . |
|---|---|---|---|---|---|---|---|---|---|---|
| Adhesion molecules | ||||||||||
| CDH1 | C-160A | rs16260 | 7493 vs 7329 (5†) | C | 0.74 | 0.72 | 0.19% | No assoc (63) | — | 1000 Genomes |
| MMP1 | G-1607GG | rs1799750 | 1007 vs 1032 (5) | G | 0.44 | 0.47 | 0.62% | Positive assoc; 1343/1590 (7) (65) | (64) | Current study |
| MMP3 | AAAAA-1612AAAAAA | rs3025058 | 857 vs 932 (4) | 5A | 0.38 | 0.41 | 0.03% | No assoc; 1296/1543 (7) (65) | (64) | Current study |
| MMP9 | 1562C/T ‡ | rs3918242 | 575 vs 836 (4) | C | — | — | — | No assoc; 575/836 (4) (64) | (64) | 1000 Genomes |
| Alcohol metabolism | ||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | His | 0.75 | 0.76 | 0.32% | n/a | — | 1000 Genomes |
| ADH1C | Ile349Val (1045A>G) | rs698 | 3168 vs 6229 (7) | Ile (A) | 0.69 | 0.64 | 0.14% | n/a | — | 1000 Genomes |
| ALDH2 | Glu487Lys | rs671 | 2209 vs 3383 (8) | Glu (G) | 0.76 | 0.74 | 0.02% | Inverse assoc; 1960/3163 (6) (66) | — | 1000 Genomes |
| Angiogenesis | ||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | C | 0.84 | 0.83 | 0.60% | n/a | — | 1000 Genomes |
| VEGF | G634C § | rs2010963 | 1508 vs 1308 (4) | C | — | — | 0.00% || | No assoc; 1508/1308 (4) (131) | — | 1000 Genomes |
| Base-excision repair | ||||||||||
| MGMT | Leu84Phe ¶ | rs12917 | 1524 vs 4646 (5) | C | 0.89 | 0.88 | 0.14% | No assoc; 1524/4646 (5) (137) | — | 1000 Genomes |
| MGMT | Il3143Val ¶ | rs2308321 | 1326 vs 3520 (4) | A | 0.89 | 0.87 | 0.19% | No assoc; 1326/3520 (4) (137) | — | 1000 Genomes |
| MUTYH | G396D | rs36053993 | 26 592 vs 19 207 (15) | G | 0.99 | 0.99 | 0.87% | Positive assoc; 25 616/18 521 (15) (48) | (138,139, (140,141) | Current study |
| MUTYH | Y179C | rs34612342 | 26 370 vs 19 042 (15) | A | 1.00 | 1.00 | 0.00% | Positive assoc; 25 392/18 362 (15) (48) | (138,139, (140,141) | Current study |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | Ser (C) | 0.71 | 0.65 | 0.02% | n/a | — | 1000 Genomes |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11†) | C | 0.91 | 0.88 | 0.16% | No assoc; 1709/3233 (9) (70) | (69) | 1000 Genomes |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | G | 0.94 | 0.95 | 0.04% | No assoc; 931/1547 (4) (70) | — | 1000 Genomes |
| XRCC1 | Arg399Gln | rs25487 | 7247 vs 8786 (12†) | G | 0.67 | 0.67 | 0.19% | No assoc; 2776/4402 (14) (70) | (69) | 1000 Genomes |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10†) | T | 0.72 | 0.71 | 0.00% | No assoc; 3183/3926 (7) (69) | — | 1000 Genomes |
| Inflammation or immune response | ||||||||||
| IL6 | 174G>C | rs1800795 | 6676 vs 7942 (10†,#) | G | 0.61 | 0.60 | 0.05% | n/a | — | 1000 Genomes |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7†) | T | 0.54 | 0.54 | 0.02% | n/a | — | 1000 Genomes |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5†) | A | 0.52 | 0.51 | 0.04% | n/a | — | 1000 Genomes |
| PPAR-ɣ | C1431T | rs3856806 | 5574 vs 7035 (7†) | C | 0.87 | 0.87 | 0.08% | No assoc; 486/941 (3) (106) | (107) | 1000 Genomes |
| PPAR-ɣ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17†,#) | C | 0.88 | 0.88 | 0.06% | Inverse assoc; 6878/9391 (10) (106) | — | 1000 Genomes |
| PTGS2/COX2 | A1195G | rs689466 | 4756 vs 6030 (7†) | A | 0.73 | 0.74 | 0.03% | Positive assoc; 1196/ 1691 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | A1803G | rs4648298 | 4229 vs 4279 (5†) | A | 0.97 | 0.97 | 0.00%** | No assoc; 480/657 (2) (142) | — | 1000 Genomes |
| PTGS2/COX2 | C427T | rs5275 | 4745 vs 5756 (7†) | T | 0.66 | 0.66 | 0.01% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G306C | rs5277 | 4269 vs 4735 (5†) | G | 0.85 | 0.84 | 0.24% | n/a | — | 1000 Genomes |
| PTGS2/COX2 | G765C | rs20417 | 5459 vs 7272 (11†) | G | 0.86 | 0.88 | 0.26% | Positive assoc; 3322/5166 (10) (123) | (142) | 1000 Genomes |
| PTGS2/COX2 | T1532C | rs5273 | 2843 vs 3216 (5†) | val/val | 1.00 | 1.00 | — | No assoc; 670/1113 (4) (142) | — | 1000 Genomes |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9†) | G | 0.80 | 0.81 | 0.02% | No assoc; 1372/1458 (7) (132) | — | Current study |
| NOD2 | 3020incC | rs5743293 | 4222 vs 2988 (8) | G | 0.96 | 0.97 | 0.95% | Positive assoc; 2571/1856 (7) (72) | — | Current study |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6†) | G | 0.99 | 0.99 | — | Positive assoc; 1442/1109 (5) (72) | — | Current study |
| NOD2 | R702W | rs2066844 | 3445 vs 2731 (6†) | C | 0.96 | 0.97 | 0.00% | Positive assoc; 1436/1109 (5) (72) | — | 1000 Genomes |
| Inhibition of cell growth | ||||||||||
| CCND1 | 870A | rs17852153 | 4747 vs 6783 (13) | G | 0.46 | 0.46 | 0.18% | Positive assoc; 2289/3232 (13) (73) | — | Current study |
| TGFB1 | C509T †† | rs1800469 | 994 vs 2335 (5) | T | 0.42 | 0.48 | 1.99% | No assoc; 994/2335 (5) (76) | — | 1000 Genomes |
| TGFBR1 | TGFBR1*6A | rs11466445 | 3217 vs 4539 (8) | C | 0.90 | 0.92 | 0.52% | Positive assoc; 5666/8450 (7) (79) | (77,78) | Current study |
| Insulin related | ||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 19/19 | 0.62 | 0.61 | 0.02% | No assoc; 3672/4125 (4) (117) | (116) | — |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | A | 0.48 | 0.50 | 0.00% | No assoc; 2834/3520 (3) (117) | — | 1000 Genomes |
| Iron metabolism | ||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6†#) | C | 0.92 | 0.93 | 0.25% | n/a | — | 1000 Genomes |
| Lipid metabolism | ||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5†) | e3 | 0.81 | 0.81 | 0.80% | n/a | — | 1000 Genomes |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5†) | e3 | 0.79 | 0.79 | 0.09% | n/a | — | 1000 Genomes |
| Mitotic control | ||||||||||
| STK15 | F31 ‡ | rs2273535 | 4860 vs 4629 (4) | T | 0.75 | 0.76 | 0.31% | Positive assoc; 2302/1769 (3) (143) | — | 1000 Genomes |
| One-carbon metabolism | ||||||||||
| MTHFR | C677T | rs1801133 | 27 372 vs 39 867 (52§) | C | 0.67 | 0.67 | 0.24% | Inverse assoc; 9345/18 887 (37) (56) | (51,52,53, 54,55,88) | 1000 Genomes |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34#) | A | 0.70 | 0.70 | 0.06% | Inverse assoc; 4764/6592 (9) (53) | — | 1000 Genomes |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14#) | A | 0.81 | 0.80 | 0.01% | No assoc; 7804/8184 (9) (110) | (88) | 1000 Genomes |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | A | 0.59 | 0.61 | 0.02% | n/a | — | 1000 Genomes |
| TS | TSER | rs34743033 | 3519 vs 5289 (5) | 3R/3R | 0.57 | 0.57 | 0.24% | n/a | — | Current study |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | ins/ins | 0.67 | 0.67 | 0.03% | n/a | — | Current study |
| Rare, high penetrance | ||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | G | 0.99 | 0.99 | — | No assoc; 3794/4484 (8) (122) | — | Current study |
| APC | D1822V | rs459552 | 6282 vs 7038 (6) | Asp | 0.78 | 0.77 | 0.38% | n/a | — | 1000 Genomes |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7†) | A | 0.71 | 0.71 | 0.02% | n/a | — | 1000 Genomes |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6†) | G | 0.77 | 0.78 | 0.12% | n/a | — | 1000 Genomes |
| Substrate metabolism | ||||||||||
| CYP1A1 | 2454A>G | rs1048943 | 10 274 vs 11 978 (13†,#) | A | 0.91 | 0.92 | 0.18% | Positive assoc; 5336/6226 (13) (87) | (88) | 1000 Genomes |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | T | 0.84 | 0.83 | 0.20% | No assoc; 234/250 (2) (88) | — | 1000 Genomes |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | A | 0.68 | 0.68 | 0.00% | n/a | — | 1000 Genomes |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6†) | C | 0.53 | 0.53 | 0.00% | n/a | — | 1000 Genomes |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6†) | C | 0.86 | 0.86 | 0.95% | n/a | — | HapMap |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6†) | A | 0.93 | 0.94 | 1.85% | n/a | — | 1000 Genomes |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8#) | C | 0.90 | 0.88 | 0.64% | n/a | — | 1000 Genomes |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | G | 0.94 | 0.93 | 1.06% | No assoc; 4979/6012 (10) (133) | — | 1000 Genomes |
| GSTA1 | GSTA1*B allele ‡‡ | — | 1648 vs 2039 (4) | A | — | — | — | No assoc; 1648/2039 (4) (92) | — | — |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | present | 0.48 | 0.49 | — | Positive assoc; 11 998/17 552 (44) (92) | (93,91,89,90,88) | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22†) | IA | 0.71 | 0.72 | 0.09% | No assoc; 5421/7671 (19) (92) | (111,88) | 1000 Genomes |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6†,#) | C | 0.92 | 0.92 | 0.22% | n/a | — | 1000 Genomes |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | present | 0.65 | 0.68 | — | Positive assoc; 8596/13 589 (34) (92) | (88,95,96) | — |
| NAT1 | slow/rapid | n/a | 4791 vs 6628 (15) | slow | 0.69 | 0.68 | 0.12% | No assoc; 520/433 (3) (88) | — | — |
| NAT2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | slow | 0.67 | 0.66 | 0.04% | No assoc; 6741/8015 (18) (88) | (108,51,109) | — |
| NQO1 | Pro187Ser (C609T) | rs1800566 | 5084 vs 5932 (8) | C | 0.81 | 0.79 | 0.00% | Positive assoc; 1783/2494 (6) (130) | — | 1000 Genomes |
| Tumor suppressor genes | ||||||||||
| TP53 | Arg72Pro §§ | rs1042522 | 7414 vs 9872 (27) | G | — | — | 0.01% | No assoc; 7414/9872 (27) (119) | (118,120,121) | 1000 Genomes |
| TP53 | intron 3 16bp |||| | rs17878362 | 1637 vs 1874 (5) | Del | — | — | 0.00% | No assoc; 1637/1874 (5) (144) | — | — |
| MDM2 | 309 T/G ¶¶ | rs2279744 | 2543 vs 2115 (7) | G | 0.46 | 0.43 | 0.42% | No assoc; 2543/2115 (7) (145) | (146) | 1000 Genomes |
| Vitamin D and calcium metabolism | ||||||||||
| VDR | BsmI (60890GA) | rs1544410 | 5607 vs 6202 (7) | G | 0.63 | 0.60 | 0.12%## | Inverse assoc; 3285/1497 (4) (147) | — | 1000 Genomes |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9#) | C | 0.61 | 0.60 | 0.00% | No assoc; 1331/2943 (5) (147) | — | 1000 Genomes |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | T | 0.68 | 0.70 | 0.01% | n/a | — | 1000 Genomes |
| Common low penetrance | ||||||||||
| SMAD7 | rs4939827 | rs4939827 | 37 650 vs 36 154 (13#) | T | 0.55 | 0.51 | 0.64% | n/a | — | 1000 Genomes |
| SMAD7 | rs12953717 | rs12953717 | 33 771 vs 32 364 (11#) | C | 0.63 | 0.65 | 0.30% | n/a | — | 1000 Genomes |
| SMAD7 | rs4464148 | rs4464148 | 15 999 vs 15 216 (7†) | T | 0.62 | 0.66 | 0.47% | n/a | — | 1000 Genomes |
| 8q24 | rs6983267 | rs6983267 | 40 604 vs 42 672 (19) | A | 0.48 | 0.51 | 1.06% | Positive assoc; (17) (148) | — | 1000 Genomes |
| 8q24 | rs10505477 | rs10505477 | 18 580 vs 20 147 (14) | C | 0.46 | 0.49 | 0.59% | n/a | — | 1000 Genomes |
| 9p24 | rs719725 | rs719725 | 13 290 vs 14 774 (13) | C | 0.37 | 0.39 | 0.16% | Positive assoc; 14 064/15 933 (17) (102) | — | 1000 Genomes |
| 19q13.1 | rs10411210 | rs10411210 | 25 607 vs 26 477 (17) | C | 0.89 | 0.88 | 0.09% | n/a | — | 1000 Genomes |
| 16q22.1 | rs9929218 | rs9929218 | 26 191 vs 27 409 (18) | G | 0.74 | 0.72 | 0.23% | n/a | — | 1000 Genomes |
| 15q14 | rs4779584 | rs4779584 | 13 656 vs 12 635 (9) | C | 0.64 | 0.65 | 0.84% | n/a | — | 1000 Genomes |
| 1q41 | rs6691170 | rs6691170 | 17 740 vs 19 776 (11) | G | 0.62 | 0.64 | 0.18% | n/a | — | 1000 Genomes |
| 3q26.2 | rs10936599 | rs10936599 | 17 802 vs 19 795 (11) | C | 0.77 | 0.75 | 0.12% | n/a | — | 1000 Genomes |
| 12q13.13 | rs11169552 | rs11169552 | 17 148 vs 19 739 (11) | C | 0.74 | 0.72 | 0.76% | n/a | — | 1000 Genomes |
| 20q13.33 | rs4925386 | rs4925386 | 17 847 vs 19 832 (11) | C | 0.71 | 0.68 | 0.39% | n/a | — | 1000 Genomes |
| 14q22.2 | rs4444235 | rs4444235 | 18 607 vs 19 576 (13) | T | 0.53 | 0.55 | 0.21% | n/a | — | 1000 Genomes |
| 20p12.3 | rs961253 | rs961253 | 18 118 vs 19 006 (13) | C | 0.66 | 0.68 | 0.22% | n/a | — | 1000 Genomes |
| 8q23.3 | rs16892766 | rs16892766 | 17 180 vs 17 840 (4†) | A | 0.88 | 0.90 | 0.10% | n/a | — | 1000 Genomes |
| 10p14 | rs10795668 | rs10795668 | 20 026 vs 20 682 (6†) | G | 0.72 | 0.69 | 0.52% | n/a | — | 1000 Genomes |
| 11q23.1 | rs3802842 | rs3802842 | 33 004 vs 31 654 (14) | A | 0.67 | 0.70 | 0.37% | n/a | — | 1000 Genomes |
* n/a = not applicable. See Supplementary Table 1 (available online) for gene names.
† Includes unpublished data from SOCCS.
‡ McColgan and Sharma 2009 (64).
§ Liu et al. 2011 (131).
|| ref allele frequency taken from 1000 Genomes data.
¶ Zhong et al. 2010 (137).
# Includes unpublished data from Ontario.
** OR for homozygote estimated as square of OR for heterozygotes.
†† Fang et al. 2010 (76).
‡‡ Economopoulos and Sergentanis 2010 (92).
§§ Economopoulos et al. 2010 (119).
|||| Hu et al. 2010 (144).
¶¶ Tomlinson 2008 was based on 10 samples.
## Based on the white-only analysis.
Statistical Analysis
Meta-analyses were conducted for 92 polymorphisms in 64 different loci (Table 1) with genotypes available in four or more candidate or GWA studies. On average, these meta-analyses were based on data from 5281 case patients (median interquartile range = 13 472 – 3384) and 6594 control subjects (median interquartile range = 16 102 – 4534) originating from eight (median interquartile range = 13 – 6) case–control samples. Unpublished data from a Scottish and/or a Canadian GWAS were included in the analyses of 37 SNPs. Summary crude odds ratios (ORs) and 95% confidence intervals for two additive models (var/wt vs wt/wt and var/var vs wt/wt) for variants that were identified from candidate studies are presented in Table 2 and for variants that were identified from GWAS, in Table 3. Summary crude odds ratios and 95% confidence intervals for a recessive (var/var vs var/wt and wt/wt) and a dominant model (var/var and var/wt vs wt/wt) are presented in Supplementary Table 2 (available online), for variants that were identified from candidate studies, and Supplementary Table 3 (available online), for variants that were identified from GWAS. For 37 associations in which the 95% confidence intervals excluded unity, we repeated the analysis excluding studies in nonwhite populations (Supplementary Tables 4 and 5, available online). We checked the linkage disequilibrium (LD) for all the polymorphisms that were included in our study and we found that the following variants were in LD (r2 threshold ≥ 0.60): 1) rs4939827 (in SMAD7) and rs12953717 (in SMAD7, r2 = 0.60); 2) rs3813867 (in CYP2E1) and rs2031920 (in CYP2E1, r2 = 0.87); 3) rs10505477 (in 8q24) and rs6983267 (in 8q24, r2 = 0.88); 4) rs731236 (in VDR) and rs1544410 (in VDR, r2 = 1.00).
Summary crude odds ratios (ORs) and 95% confidence intervals (95% CIs) for two additive models for variants that were identified from candidate studies
| Gene . | Variant . | rs number . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | I2(95% CI) . | Power . | N . | OR (95% CI) . | P . | I2(95% CI) . | Power . | BFDP* . | Venice criteria grade† . | N . | ||||
| Adhesion molecules | |||||||||||||||
| CDH1 | C-160A‡ | rs16260 | 7493 vs 7329 (5§) | 5 | 0.91 (0.85 to 0.97) | .005 | 49 (0 to 81) | 0.78 | 5 | 0.84 (0.74 to 0.96) | .01 | 34 (0 to 75) | 0.76 | 0.67 | BBB |
| MMP1 | G-1607GG‡ | rs1799750 | 1007 vs 1032 (5) | 5 | 1.05 (0.84 to 1.33) | .66 | 15 (0 to 82) | 0.07 | 5 | 1.54 (1.00 to 2.37) | .05 | 53 (0 to 83) | 0.61 | 0.88 | BBB |
| MMP3 | AAAAA-612AAAAAA‡ | rs3025058 | 857 vs 932 (4) | 4 | 0.79 (0.60 to 1.03) | .08 | 0 (0 to 85) | 0.43 | 4 | 1.16 (0.86 to 1.56) | .33 | 0 (0 to 85) | 0.20 | 0.92 | CAB |
| MMP9 | 1562C/T|| | rs3918242 | 575 vs 836 (4) | — | — | — | — | — | — | — | — | — | — | — | — |
| Alcohol metabolism | |||||||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | 5 | 1.06 (0.83 to 1.36) | .63 | 70 (24 to 88) | 0.15 | 5 | 1.22 (0.94 to 1.58) | .13 | 26 (0 to 70) | 0.35 | 0.97 | CCB |
| ADH1C | Ile349Val(1045A>G) | rs698 | 3168 vs 6229 (7) | 7 | 0.96 (0.87 to 1.07) | .49 | 0 (0 to 78) | 0.12 | 7 | 0.88 (0.66 to 1.16) | .35 | 65 (21 to 84) | 0.4 | 0.98 | CBB |
| ALDH2 | Glu487Lys | rs671‡ | 2209 vs 3383 (8) | 8 | 0.88 (0.79 to 0.99) | .04 | 0 (0 to 65) | 0.59 | 8 | 0.89 (0.58 to 1.36) | .59 | 58 (12 to 80) | 0.17 | 0.89 | CCB |
| Angiogenesis | |||||||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | 4 | 0.94 (0.71 to 1.24) | .69 | 55 (0 to 85) | 0.13 | 4 | 1.19 (0.72 to 1.99) | .50 | 15 (0 to 87) | 0.10 | 0.97 | CBB |
| VEGF | G634C¶ | rs2010963 | 1508 vs 1308 (4) | 4 | 0.89 (0.72 to 1.10) | — | — | — | 4 | 1.17 (0.93 to 1.47) | — | — | — | 0.96 | --C |
| Base-excision repair | |||||||||||||||
| MGMT | Leu84Phe** | rs12917 | 1524 vs 4646 (5) | 5 | 0.84 (0.70 to 1.00) | .05 | — | — | 5 | 0.97 (0.56 to 1.66) | .97 | — | — | 0.91 | --C |
| MGMT | Il3143Val** | rs2308321 | 1326 vs 3520 (4) | 4 | 0.86 (0.66 to 1.12) | .26 | — | — | 4 | 1.01 (0.56 to 1.81) | .69 | — | — | 0.91 | --C |
| MUTYH | G396D | rs36053993†† | 26 592 vs 19 207 (15) | 15 | 1.07 (0.91 to 1.27) | .41 | 0 (0 to 54) | 0.13 | 9 | 6.15 (2.34 to 16.15) | .00 | 0 (0 to 65) | 0.50 | 0.17 | CAB |
| MUTYH | Y179C | rs34612342‡ | 26 370 vs 19 042 (15) | 14 | 1.34 (1.02 to 1.77) | .04 | 0 (0 to 55) | 0.55 | 6 | 3.35 (1.14 to 9.89) | .03 | 0 (0 to 75) | 0.37 | 0.89 | BAB |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | 9 | 1.02 (0.94 to 1.12) | .60 | 48 (0 to 76) | 0.08 | 9 | 1.05 (0.89 to 1.23) | .58 | 42 (0 to 73) | 0.14 | 0.99 | CBB |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11§) | 10 | 0.96 (0.87 to 1.07) | .50 | 0 (0 to 62) | 0.13 | 10 | 1.10 (0.82 to 1.48) | .52 | 16 (0 to 57) | 0.13 | 0.98 | CAB |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | 4 | 1.06 (0.83 to 1.34) | .65 | 41 (0 to 80) | 0.08 | 3 | 1.16 (0.37 to 3.62) | .31 | 14 (0 to 91) | 0.06 | 0.97 | CBB |
| XRCC1 | Arg399Gln | rs25487‡ | 7247 vs 8786 (12§) | 12 | 0.99 (0.92 to 1.06) | .72 | 11 (0 to 51) | 0.06 | 12 | 0.88 (0.79 to 0.97) | .02 | 0 (0 to 58) | 0.67 | 0.99 | BAB |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10§) | 10 | 0.92 (0.84 to 1.01) | .09 | 36 (0 to 69) | 0.48 | 10 | 0.95 (0.72 to 1.24) | .68 | 57 (12 to 79) | 0.12 | 0.95 | CBB |
| Inflammation or immune response | |||||||||||||||
| IL6 | 174G>C | rs1800795‡ | 6676 vs 7942 (10§,||) | 10 | 1.03 (0.91 to 1.17) | .65 | 56 (11 to 78) | 0.13 | 10 | 0.94 (0.79 to 1.12) | .48 | 56 (11 to 78) | 0.24 | 0.98 | CCB |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7§) | 7 | 1.03 (0.92 to 1.15) | .61 | 44 (0 to 77) | 0.08 | 7 | 1.05 (0.91 to 1.20) | .53 | 0 (0 to 71) | 0.11 | 0.99 | CBB |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5§) | 5 | 0.96 (0.85 to 1.08) | .49 | 0 (0 to 79) | 0.11 | 5 | 0.93 (0.81 to 1.07) | .30 | 35 (0 to 75) | 0.18 | 0.98 | CAB |
| PPARγ | C1431T | rs3856806 | 5574 vs 7035 (7§) | 5 | 1.04 (0.95 to 1.13) | .44 | 0 (0 to 79) | 0.14 | 5 | 0.95 (0.50 to 1.78) | .86 | 69 (22 to 88) | 0.06 | 0.98 | CAB |
| PPARγ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17§,||) | 13 | 0.98 (0.86 to 1.11) | .72 | 65 (36 to 80) | 0.09 | 12 | 0.91 (0.73 to 1.12) | .37 | 0 (0 to 58) | 0.14 | 0.98 | CCB |
| PTGS2 | A1195G | rs689466 | 4756 vs 6030 (7§) | 7 | 1.04 (0.95 to 1.13) | .42 | 41 (0 to 75) | 0.16 | 7 | 1.08 (0.77 to 1.51) | .66 | 64 (19 to 84) | 0.19 | 0.98 | CBB |
| PTGS2 | A1803G | rs4648298 | 4229 vs 4279 (5§) | 3 | 1.00 (0.83 to 1.22) | .96 | 49 (0 to 85) | 0.05 | 3 | n/a | n/a | n/a | n/a | 0.98 | CBB |
| PTGS2 | C427T | rs5275 | 4745 vs 5756 (7§) | 7 | 1.01 (0.93 to 1.09) | .87 | 0 (0 to 71) | 0.06 | 7 | 1.03 (0.91 to 1.17) | .65 | 0 (0 to 71) | 0.08 | 0.99 | CAB |
| PTGS2 | G306C | rs5277 | 4269 vs 4735 (5§) | 5 | 0.97 (0.88 to 1.06) | .45 | 23 (0 to 68) | 0.10 | 5 | 0.85 (0.65 to 1.11) | .24 | 5 (0 to 80) | 0.23 | 0.99 | CAB |
| PTGS2 | G765C | rs20417 | 5459 vs 7272 (11§) | 10 | 1.03 (0.95 to 1.13) | .45 | 45 (0 to 74) | 0.11 | 8 | 1.21 (0.93 to 1.57) | .15 | 0 (0 to 68) | 0.3 | 0.99 | CBB |
| PTGS2 | T1532C | rs5273 | 2843 vs 3216 (5§) | 2 | n/a | n/a | n/a | n/a | 2 | n/a | n/a | n/a | n/a | — | — |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9§) | 9 | 1.11 (0.88 to 1.40) | .37 | 72 (45 to 86) | 0.65 | 9 | 1.13 (0.92 to 1.39) | .24 | 47 (0 to 75) | 0.21 | 0.97 | BCB |
| NOD2 | 3020incC‡ | rs5743293 | 4222 vs 2988 (8) | 8 | 1.39 (1.15 to 1.69) | .001 | 0 (0 to 68) | 0.94 | 5 | 2.81 (0.87 to 9.05) | .08 | 0 (0 to 79) | 0.26 | 0.34 | AAB |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6§) | 6 | 1.41 (1.04 to 1.91) | .03 | 0 (0 to 75) | 0.63 | 0 | n/a | n/a | n/a | n/a | 0.87 | BBB |
| NOD2 | R702W‡ | rs2066844 | 3445 vs 2731 (6§) | 6 | 1.22 (1.00 to 1.50) | .06 | 12 (0 to 78) | 0.49 | 4 | 1.23 (0.41 to 3.70) | .71 | 0 (0 to 85) | 0.06 | 0.91 | BAB |
| Inhibition of cell growth | |||||||||||||||
| CCND1 | 870A‡ | rs17852153 | 4747 vs 6783 (13) | 13 | 1.13 (1.03 to 1.25) | .02 | 0 (0 to 57) | 0.70 | 13 | 1.16 (0.98 to 1.38) | .09 | 51 (8 to 74) | 0.78 | 0.85 | BAB |
| TGFB1 | C509T‡‡ | rs1800469 | 994 vs 2335 (5) | 5 | 1.12 (0.91 to 1.37) | — | — | — | 5 | 1.62 (1.30 to 2.02) | — | — | — | 0.96 | --C |
| TGFBR1 | TGFBR1*6A‡ | rs11466445 | 3217 vs 4539 (8) | 8 | 1.15 (1.01 to 1.31) | .03 | 0 (0 to 68) | — | 8 | 1.51 (0.69 to 3.31) | .31 | 52 (0 to 87) | — | 0.89 | -AB |
| Insulin-related | |||||||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 6 | 1.06 (0.99 to 1.14) | .08 | 0 (0 to 75) | 0.4 | 6 | 1.07 (0.97 to 1.18) | .15 | 0 (0 to 75) | 0.31 | 0.97 | CAB |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | 6 | 1.02 (0.94 to 1.10) | .72 | 0 (0 to 75) | 0.08 | 6 | 1.00 (0.91 to 1.10) | .96 | 0 (0 to 75) | 0.05 | 0.99 | CAB |
| Iron metabolism | |||||||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6§,||) | 6 | 1.08 (0.97 to 1.21) | .15 | 0 (0 to 75) | 0.29 | 5 | 0.90 (0.55 to 1.48) | .69 | 0 (0 to 79) | 0.07 | 0.97 | CAB |
| Lipid metabolism | |||||||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5§) | 5 | 0.94 (0.85 to 1.04) | .23 | 66 (11 to 87) | 0.21 | 4 | 0.75 (0.46 to 1.22) | .25 | 0 (0 to 85) | 0.21 | 0.98 | CCB |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5§) | 5 | 1.04 (0.94 to 1.15) | .49 | 14 (0 to 82) | 0.12 | 5 | 1.12 (0.83 to 1.52) | .45 | 0 (0 to 79) | 0.12 | 0.98 | CAB |
| Mitotic control | |||||||||||||||
| STK15 | F31I | rs2273535 | 4860 vs 4629 (4) | 4 | 1.03 (0.94 to 1.12) | .58 | 0 (0 to 85) | 0.1 | 4 | 1.30 (0.96 to 1.76) | .09 | 52 (0 to 84) | 0.56 | 0.99 | CAB |
| One-carbon metabolism | |||||||||||||||
| MTHFR | C677T | rs1801133†† | 27 372 vs 39 867 (52||) | 52 | 1.00 (0.94 to 1.06) | .92 | 53 (36 to 65) | 0.05 | 52 | 0.87 (0.82 to 0.91) | .00 | 35 (10 to 53) | 1.00 | 0.99 | CBB |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34||) | 34 | 1.01 (0.97 to 1.06) | .51 | 0 (0 to 37) | 0.08 | 34 | 0.94 (0.87 to 1.01) | .09 | 22 (0 to 49) | 0.40 | 0.99 | CAB |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14||) | 14 | 0.97 (0.92 to 1.02) | .27 | 12 (0 to 50) | 0.21 | 14 | 0.96 (0.84 to 1.09) | .50 | 48 (4 to 72) | 0.05 | 0.99 | CAB |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | 9 | 0.98 (0.90 to 1.07) | .66 | 15 (0 to 57) | 0.10 | 9 | 1.04 (0.94 to 1.14) | .47 | 23 (0 to 64) | 0.17 | 0.99 | CAB |
| TS | TSER‡ | rs34743033 | 3519 vs 5289 (5) | 5 | 0.86 (0.78 to 0.95) | .003 | 18 (0 to 83) | 0.87 | 5 | 0.83 (0.73 to 0.94) | .004 | 17 (0 to 83) | 0.85 | 0.55 | AAB |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | 4 | 0.96 (0.88 to 1.06) | .45 | 0 (0 to 85) | 0.13 | 4 | 1.03 (0.88 to 1.19) | .73 | 0 (0 to 85) | 0.07 | 0.98 | CAB |
| Rare, high penetrance | |||||||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | 6 | 1.13 (0.88 to 1.47) | .34 | 0 (0 to 75) | 0.15 | 1 | n/a | n/a | n/a | n/a | 0.96 | CAB |
| APC | D1822V‡ | rs459552 | 6282 vs 7038 (6) | 6 | 0.99 (0.92 to 1.07) | .83 | 24 (0 to 68) | 0.06 | 6 | 0.84 (0.71 to 0.98) | .03 | 0 (0 to 75) | 0.55 | 0.99 | CAB |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7§) | 7 | 1.09 (0.90 to 1.32) | .40 | 55 (0 to 81) | 0.42 | 7 | 1.01 (0.85 to 1.21) | .88 | 43 (0 to 76) | 0.05 | 0.97 | CCB |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6§) | 5 | 1.06 (0.97 to 1.15) | .23 | 0 (0 to 79) | 0.27 | 5 | 1.15 (0.95 to 1.39) | .15 | 26 (0 to 71) | 0.33 | 0.97 | CAB |
| Substrate metabolism | |||||||||||||||
| CYP1A1 | 2454A>G‡ | rs1048943 | 10 274 vs 11 978 (13§,||) | 13 | 1.28 (1.01 to 1.63) | .05 | 85 (7 to 91) | 1.00 | 12 | 1.47 (1.17 to 1.85) | .001 | 3 (0 to 60) | 0.93 | 0.89 | ACB |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | 7 | 0.94 (0.86 to 1.04) | .23 | 0 (0 to 71) | 0.27 | 7 | 0.84 (0.56 to 1.27) | .42 | 54 (0 to 80) | 0.38 | 0.98 | CAB |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | 9 | 1.13 (0.95 to 1.34) | .18 | 63 (25 to 82) | 0.75 | 9 | 1.07 (0.92 to 1.26) | .40 | 42 (0 to 73) | 0.16 | 0.96 | ACB |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6§) | 6 | 0.99 (0.92 to 1.06) | .69 | 0 (0 to 75) | 0.06 | 6 | 0.98 (0.90 to 1.07) | .70 | 0 (0 to 77) | 0.08 | 0.99 | CAB |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6§) | 6 | 0.93 (0.85 to 1.02) | .13 | 21 (0 to 65) | 0.34 | 6 | 1.29 (0.99 to 1.70) | .06 | 0 (0 to 75) | 0.46 | 0.97 | CAB |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6§) | 6 | 1.08 (0.83 to 1.40) | .57 | 73 (33 to 88) | 0.26 | 4 | 0.68 (0.37 to 1.26) | .22 | 11 (0 to 86) | 0.23 | 0.97 | CCB |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8||) | 8 | 0.93 (0.83 to 1.05) | .23 | 0 (0 to 68) | 0.27 | 8 | 1.23 (0.92 to 1.63) | .16 | 35 (0 to 71) | 0.34 | 0.97 | CAB |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | 7 | 1.17 (0.92 to 1.48) | .21 | 53 (0 to 80) | 0.61 | 6 | 1.83 (0.94 to 3.57) | .08 | 0 (0 to 75) | 0.55 | 0.95 | BCB |
| GSTA1 | GSTA1*B§§ | 1648 vs 2039 (4) | 4 | 1.03 (0.89 to 1.19) | — | — | — | 4 | 1.09 (0.90 to 1.32) | — | — | — | 0.98 | — | |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22§) | 22 | 1.05 (0.99 to 1.12) | .11 | 0 (0 to 46) | 0.38 | 22 | 0.95 (0.86 to 1.05) | .32 | 36 (0 to 62) | 0.17 | 0.98 | CAB |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6§,||) | 6 | 1.02 (0.91 to 1.13) | .77 | 0 (0 to 75) | 0.07 | 6 | 0.87 (0.55 to 1.37) | .55 | 12 (0 to 78) | 0.09 | 0.99 | CAB |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| ΝΑΤ1 | slow/rapid‡ | n/a | 4791 vs 6628 (15) | 7 | 0.80 (0.68 to 0.93) | .003 | 38 (0 to 74) | 0.86 | 7 | 0.98 0.79 to 1.22) | .97 | 0 (0 to 71) | 0.06 | 0.57 | ABB |
| ΝΑΤ2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | 15 | 1.01 (0.83 to 1.22) | .94 | 81 (70 to 88) | 0.06 | 15 | 0.95 (0.76 to 1.20) | .68 | 64 (37 to 79) | 0.17 | 0.98 | CCB |
| NQO1 | Pro187Ser(C609T) | rs1800566 | 5084 vs 5932 (8) | 8 | 1.14 (0.96 to 1.35) | .12 | 64 (22 to 83) | 0.89 | 8 | 1.10 (0.76 to 1.59) | .63 | 56 (2 to 80) | 0.19 | 0.95 | ACB |
| Tumor suppressor genes | |||||||||||||||
| TP53 | Arg72Pro|||| | rs1042522 | 7414 vs 9872 (27) | 27 | 1.01 (0.89 to 1.14) | .90 | — | — | 27 | 1.04 (0.82 to 1.31) | .77 | — | — | 0.99 | --C |
| TP53 | intron3 16bp|||| | rs17878362 | 1637 vs 1874 (5) | 5 | 1.58 (0.98 to 2.56) | — | — | — | 5 | 1.14 (0.84 to 1.55) | — | — | — | 0.92 | --C |
| MDM2 | 309 T/G|||| | rs2279744 | 2543 vs 2115 (7) | 7 | 0.73 (0.62 to 0.86) | — | — | — | 7 | 0.86 (0.57 to 1.30) | — | — | — | 0.10 | --C |
| Vitamin D and calcium metabolism | |||||||||||||||
| VDR | BsmI(60890GA) | rs1544410‡ | 5607 vs 6202 (7) | 7 | 0.77 (0.58 to 1.02) | .07 | 89 (81 to 94) | 1.00 | 7 | 0.51 (0.28 to 0.90) | .02 | 95 (92 to 97) | 1.00 | 0.92 | ACB |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9||) | 9 | 0.97 (0.85 to 1.11) | .66 | 67 (32 to 83) | 0.16 | 9 | 0.99 (0.82 to 1.20) | .94 | 70 (41 to 85) | 0.06 | 0.98 | CCB |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | 4 | 1.06 (0.87 to 1.30) | .55 | 0 (0 to 85) | 0.09 | 3 | 1.00 (0.57 to 1.75) | .99 | 73 (8 to 92) | 0.05 | 0.97 | CAB |
| Gene . | Variant . | rs number . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | I2(95% CI) . | Power . | N . | OR (95% CI) . | P . | I2(95% CI) . | Power . | BFDP* . | Venice criteria grade† . | N . | ||||
| Adhesion molecules | |||||||||||||||
| CDH1 | C-160A‡ | rs16260 | 7493 vs 7329 (5§) | 5 | 0.91 (0.85 to 0.97) | .005 | 49 (0 to 81) | 0.78 | 5 | 0.84 (0.74 to 0.96) | .01 | 34 (0 to 75) | 0.76 | 0.67 | BBB |
| MMP1 | G-1607GG‡ | rs1799750 | 1007 vs 1032 (5) | 5 | 1.05 (0.84 to 1.33) | .66 | 15 (0 to 82) | 0.07 | 5 | 1.54 (1.00 to 2.37) | .05 | 53 (0 to 83) | 0.61 | 0.88 | BBB |
| MMP3 | AAAAA-612AAAAAA‡ | rs3025058 | 857 vs 932 (4) | 4 | 0.79 (0.60 to 1.03) | .08 | 0 (0 to 85) | 0.43 | 4 | 1.16 (0.86 to 1.56) | .33 | 0 (0 to 85) | 0.20 | 0.92 | CAB |
| MMP9 | 1562C/T|| | rs3918242 | 575 vs 836 (4) | — | — | — | — | — | — | — | — | — | — | — | — |
| Alcohol metabolism | |||||||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | 5 | 1.06 (0.83 to 1.36) | .63 | 70 (24 to 88) | 0.15 | 5 | 1.22 (0.94 to 1.58) | .13 | 26 (0 to 70) | 0.35 | 0.97 | CCB |
| ADH1C | Ile349Val(1045A>G) | rs698 | 3168 vs 6229 (7) | 7 | 0.96 (0.87 to 1.07) | .49 | 0 (0 to 78) | 0.12 | 7 | 0.88 (0.66 to 1.16) | .35 | 65 (21 to 84) | 0.4 | 0.98 | CBB |
| ALDH2 | Glu487Lys | rs671‡ | 2209 vs 3383 (8) | 8 | 0.88 (0.79 to 0.99) | .04 | 0 (0 to 65) | 0.59 | 8 | 0.89 (0.58 to 1.36) | .59 | 58 (12 to 80) | 0.17 | 0.89 | CCB |
| Angiogenesis | |||||||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | 4 | 0.94 (0.71 to 1.24) | .69 | 55 (0 to 85) | 0.13 | 4 | 1.19 (0.72 to 1.99) | .50 | 15 (0 to 87) | 0.10 | 0.97 | CBB |
| VEGF | G634C¶ | rs2010963 | 1508 vs 1308 (4) | 4 | 0.89 (0.72 to 1.10) | — | — | — | 4 | 1.17 (0.93 to 1.47) | — | — | — | 0.96 | --C |
| Base-excision repair | |||||||||||||||
| MGMT | Leu84Phe** | rs12917 | 1524 vs 4646 (5) | 5 | 0.84 (0.70 to 1.00) | .05 | — | — | 5 | 0.97 (0.56 to 1.66) | .97 | — | — | 0.91 | --C |
| MGMT | Il3143Val** | rs2308321 | 1326 vs 3520 (4) | 4 | 0.86 (0.66 to 1.12) | .26 | — | — | 4 | 1.01 (0.56 to 1.81) | .69 | — | — | 0.91 | --C |
| MUTYH | G396D | rs36053993†† | 26 592 vs 19 207 (15) | 15 | 1.07 (0.91 to 1.27) | .41 | 0 (0 to 54) | 0.13 | 9 | 6.15 (2.34 to 16.15) | .00 | 0 (0 to 65) | 0.50 | 0.17 | CAB |
| MUTYH | Y179C | rs34612342‡ | 26 370 vs 19 042 (15) | 14 | 1.34 (1.02 to 1.77) | .04 | 0 (0 to 55) | 0.55 | 6 | 3.35 (1.14 to 9.89) | .03 | 0 (0 to 75) | 0.37 | 0.89 | BAB |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | 9 | 1.02 (0.94 to 1.12) | .60 | 48 (0 to 76) | 0.08 | 9 | 1.05 (0.89 to 1.23) | .58 | 42 (0 to 73) | 0.14 | 0.99 | CBB |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11§) | 10 | 0.96 (0.87 to 1.07) | .50 | 0 (0 to 62) | 0.13 | 10 | 1.10 (0.82 to 1.48) | .52 | 16 (0 to 57) | 0.13 | 0.98 | CAB |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | 4 | 1.06 (0.83 to 1.34) | .65 | 41 (0 to 80) | 0.08 | 3 | 1.16 (0.37 to 3.62) | .31 | 14 (0 to 91) | 0.06 | 0.97 | CBB |
| XRCC1 | Arg399Gln | rs25487‡ | 7247 vs 8786 (12§) | 12 | 0.99 (0.92 to 1.06) | .72 | 11 (0 to 51) | 0.06 | 12 | 0.88 (0.79 to 0.97) | .02 | 0 (0 to 58) | 0.67 | 0.99 | BAB |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10§) | 10 | 0.92 (0.84 to 1.01) | .09 | 36 (0 to 69) | 0.48 | 10 | 0.95 (0.72 to 1.24) | .68 | 57 (12 to 79) | 0.12 | 0.95 | CBB |
| Inflammation or immune response | |||||||||||||||
| IL6 | 174G>C | rs1800795‡ | 6676 vs 7942 (10§,||) | 10 | 1.03 (0.91 to 1.17) | .65 | 56 (11 to 78) | 0.13 | 10 | 0.94 (0.79 to 1.12) | .48 | 56 (11 to 78) | 0.24 | 0.98 | CCB |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7§) | 7 | 1.03 (0.92 to 1.15) | .61 | 44 (0 to 77) | 0.08 | 7 | 1.05 (0.91 to 1.20) | .53 | 0 (0 to 71) | 0.11 | 0.99 | CBB |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5§) | 5 | 0.96 (0.85 to 1.08) | .49 | 0 (0 to 79) | 0.11 | 5 | 0.93 (0.81 to 1.07) | .30 | 35 (0 to 75) | 0.18 | 0.98 | CAB |
| PPARγ | C1431T | rs3856806 | 5574 vs 7035 (7§) | 5 | 1.04 (0.95 to 1.13) | .44 | 0 (0 to 79) | 0.14 | 5 | 0.95 (0.50 to 1.78) | .86 | 69 (22 to 88) | 0.06 | 0.98 | CAB |
| PPARγ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17§,||) | 13 | 0.98 (0.86 to 1.11) | .72 | 65 (36 to 80) | 0.09 | 12 | 0.91 (0.73 to 1.12) | .37 | 0 (0 to 58) | 0.14 | 0.98 | CCB |
| PTGS2 | A1195G | rs689466 | 4756 vs 6030 (7§) | 7 | 1.04 (0.95 to 1.13) | .42 | 41 (0 to 75) | 0.16 | 7 | 1.08 (0.77 to 1.51) | .66 | 64 (19 to 84) | 0.19 | 0.98 | CBB |
| PTGS2 | A1803G | rs4648298 | 4229 vs 4279 (5§) | 3 | 1.00 (0.83 to 1.22) | .96 | 49 (0 to 85) | 0.05 | 3 | n/a | n/a | n/a | n/a | 0.98 | CBB |
| PTGS2 | C427T | rs5275 | 4745 vs 5756 (7§) | 7 | 1.01 (0.93 to 1.09) | .87 | 0 (0 to 71) | 0.06 | 7 | 1.03 (0.91 to 1.17) | .65 | 0 (0 to 71) | 0.08 | 0.99 | CAB |
| PTGS2 | G306C | rs5277 | 4269 vs 4735 (5§) | 5 | 0.97 (0.88 to 1.06) | .45 | 23 (0 to 68) | 0.10 | 5 | 0.85 (0.65 to 1.11) | .24 | 5 (0 to 80) | 0.23 | 0.99 | CAB |
| PTGS2 | G765C | rs20417 | 5459 vs 7272 (11§) | 10 | 1.03 (0.95 to 1.13) | .45 | 45 (0 to 74) | 0.11 | 8 | 1.21 (0.93 to 1.57) | .15 | 0 (0 to 68) | 0.3 | 0.99 | CBB |
| PTGS2 | T1532C | rs5273 | 2843 vs 3216 (5§) | 2 | n/a | n/a | n/a | n/a | 2 | n/a | n/a | n/a | n/a | — | — |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9§) | 9 | 1.11 (0.88 to 1.40) | .37 | 72 (45 to 86) | 0.65 | 9 | 1.13 (0.92 to 1.39) | .24 | 47 (0 to 75) | 0.21 | 0.97 | BCB |
| NOD2 | 3020incC‡ | rs5743293 | 4222 vs 2988 (8) | 8 | 1.39 (1.15 to 1.69) | .001 | 0 (0 to 68) | 0.94 | 5 | 2.81 (0.87 to 9.05) | .08 | 0 (0 to 79) | 0.26 | 0.34 | AAB |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6§) | 6 | 1.41 (1.04 to 1.91) | .03 | 0 (0 to 75) | 0.63 | 0 | n/a | n/a | n/a | n/a | 0.87 | BBB |
| NOD2 | R702W‡ | rs2066844 | 3445 vs 2731 (6§) | 6 | 1.22 (1.00 to 1.50) | .06 | 12 (0 to 78) | 0.49 | 4 | 1.23 (0.41 to 3.70) | .71 | 0 (0 to 85) | 0.06 | 0.91 | BAB |
| Inhibition of cell growth | |||||||||||||||
| CCND1 | 870A‡ | rs17852153 | 4747 vs 6783 (13) | 13 | 1.13 (1.03 to 1.25) | .02 | 0 (0 to 57) | 0.70 | 13 | 1.16 (0.98 to 1.38) | .09 | 51 (8 to 74) | 0.78 | 0.85 | BAB |
| TGFB1 | C509T‡‡ | rs1800469 | 994 vs 2335 (5) | 5 | 1.12 (0.91 to 1.37) | — | — | — | 5 | 1.62 (1.30 to 2.02) | — | — | — | 0.96 | --C |
| TGFBR1 | TGFBR1*6A‡ | rs11466445 | 3217 vs 4539 (8) | 8 | 1.15 (1.01 to 1.31) | .03 | 0 (0 to 68) | — | 8 | 1.51 (0.69 to 3.31) | .31 | 52 (0 to 87) | — | 0.89 | -AB |
| Insulin-related | |||||||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 6 | 1.06 (0.99 to 1.14) | .08 | 0 (0 to 75) | 0.4 | 6 | 1.07 (0.97 to 1.18) | .15 | 0 (0 to 75) | 0.31 | 0.97 | CAB |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | 6 | 1.02 (0.94 to 1.10) | .72 | 0 (0 to 75) | 0.08 | 6 | 1.00 (0.91 to 1.10) | .96 | 0 (0 to 75) | 0.05 | 0.99 | CAB |
| Iron metabolism | |||||||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6§,||) | 6 | 1.08 (0.97 to 1.21) | .15 | 0 (0 to 75) | 0.29 | 5 | 0.90 (0.55 to 1.48) | .69 | 0 (0 to 79) | 0.07 | 0.97 | CAB |
| Lipid metabolism | |||||||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5§) | 5 | 0.94 (0.85 to 1.04) | .23 | 66 (11 to 87) | 0.21 | 4 | 0.75 (0.46 to 1.22) | .25 | 0 (0 to 85) | 0.21 | 0.98 | CCB |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5§) | 5 | 1.04 (0.94 to 1.15) | .49 | 14 (0 to 82) | 0.12 | 5 | 1.12 (0.83 to 1.52) | .45 | 0 (0 to 79) | 0.12 | 0.98 | CAB |
| Mitotic control | |||||||||||||||
| STK15 | F31I | rs2273535 | 4860 vs 4629 (4) | 4 | 1.03 (0.94 to 1.12) | .58 | 0 (0 to 85) | 0.1 | 4 | 1.30 (0.96 to 1.76) | .09 | 52 (0 to 84) | 0.56 | 0.99 | CAB |
| One-carbon metabolism | |||||||||||||||
| MTHFR | C677T | rs1801133†† | 27 372 vs 39 867 (52||) | 52 | 1.00 (0.94 to 1.06) | .92 | 53 (36 to 65) | 0.05 | 52 | 0.87 (0.82 to 0.91) | .00 | 35 (10 to 53) | 1.00 | 0.99 | CBB |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34||) | 34 | 1.01 (0.97 to 1.06) | .51 | 0 (0 to 37) | 0.08 | 34 | 0.94 (0.87 to 1.01) | .09 | 22 (0 to 49) | 0.40 | 0.99 | CAB |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14||) | 14 | 0.97 (0.92 to 1.02) | .27 | 12 (0 to 50) | 0.21 | 14 | 0.96 (0.84 to 1.09) | .50 | 48 (4 to 72) | 0.05 | 0.99 | CAB |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | 9 | 0.98 (0.90 to 1.07) | .66 | 15 (0 to 57) | 0.10 | 9 | 1.04 (0.94 to 1.14) | .47 | 23 (0 to 64) | 0.17 | 0.99 | CAB |
| TS | TSER‡ | rs34743033 | 3519 vs 5289 (5) | 5 | 0.86 (0.78 to 0.95) | .003 | 18 (0 to 83) | 0.87 | 5 | 0.83 (0.73 to 0.94) | .004 | 17 (0 to 83) | 0.85 | 0.55 | AAB |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | 4 | 0.96 (0.88 to 1.06) | .45 | 0 (0 to 85) | 0.13 | 4 | 1.03 (0.88 to 1.19) | .73 | 0 (0 to 85) | 0.07 | 0.98 | CAB |
| Rare, high penetrance | |||||||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | 6 | 1.13 (0.88 to 1.47) | .34 | 0 (0 to 75) | 0.15 | 1 | n/a | n/a | n/a | n/a | 0.96 | CAB |
| APC | D1822V‡ | rs459552 | 6282 vs 7038 (6) | 6 | 0.99 (0.92 to 1.07) | .83 | 24 (0 to 68) | 0.06 | 6 | 0.84 (0.71 to 0.98) | .03 | 0 (0 to 75) | 0.55 | 0.99 | CAB |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7§) | 7 | 1.09 (0.90 to 1.32) | .40 | 55 (0 to 81) | 0.42 | 7 | 1.01 (0.85 to 1.21) | .88 | 43 (0 to 76) | 0.05 | 0.97 | CCB |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6§) | 5 | 1.06 (0.97 to 1.15) | .23 | 0 (0 to 79) | 0.27 | 5 | 1.15 (0.95 to 1.39) | .15 | 26 (0 to 71) | 0.33 | 0.97 | CAB |
| Substrate metabolism | |||||||||||||||
| CYP1A1 | 2454A>G‡ | rs1048943 | 10 274 vs 11 978 (13§,||) | 13 | 1.28 (1.01 to 1.63) | .05 | 85 (7 to 91) | 1.00 | 12 | 1.47 (1.17 to 1.85) | .001 | 3 (0 to 60) | 0.93 | 0.89 | ACB |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | 7 | 0.94 (0.86 to 1.04) | .23 | 0 (0 to 71) | 0.27 | 7 | 0.84 (0.56 to 1.27) | .42 | 54 (0 to 80) | 0.38 | 0.98 | CAB |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | 9 | 1.13 (0.95 to 1.34) | .18 | 63 (25 to 82) | 0.75 | 9 | 1.07 (0.92 to 1.26) | .40 | 42 (0 to 73) | 0.16 | 0.96 | ACB |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6§) | 6 | 0.99 (0.92 to 1.06) | .69 | 0 (0 to 75) | 0.06 | 6 | 0.98 (0.90 to 1.07) | .70 | 0 (0 to 77) | 0.08 | 0.99 | CAB |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6§) | 6 | 0.93 (0.85 to 1.02) | .13 | 21 (0 to 65) | 0.34 | 6 | 1.29 (0.99 to 1.70) | .06 | 0 (0 to 75) | 0.46 | 0.97 | CAB |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6§) | 6 | 1.08 (0.83 to 1.40) | .57 | 73 (33 to 88) | 0.26 | 4 | 0.68 (0.37 to 1.26) | .22 | 11 (0 to 86) | 0.23 | 0.97 | CCB |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8||) | 8 | 0.93 (0.83 to 1.05) | .23 | 0 (0 to 68) | 0.27 | 8 | 1.23 (0.92 to 1.63) | .16 | 35 (0 to 71) | 0.34 | 0.97 | CAB |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | 7 | 1.17 (0.92 to 1.48) | .21 | 53 (0 to 80) | 0.61 | 6 | 1.83 (0.94 to 3.57) | .08 | 0 (0 to 75) | 0.55 | 0.95 | BCB |
| GSTA1 | GSTA1*B§§ | 1648 vs 2039 (4) | 4 | 1.03 (0.89 to 1.19) | — | — | — | 4 | 1.09 (0.90 to 1.32) | — | — | — | 0.98 | — | |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22§) | 22 | 1.05 (0.99 to 1.12) | .11 | 0 (0 to 46) | 0.38 | 22 | 0.95 (0.86 to 1.05) | .32 | 36 (0 to 62) | 0.17 | 0.98 | CAB |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6§,||) | 6 | 1.02 (0.91 to 1.13) | .77 | 0 (0 to 75) | 0.07 | 6 | 0.87 (0.55 to 1.37) | .55 | 12 (0 to 78) | 0.09 | 0.99 | CAB |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| ΝΑΤ1 | slow/rapid‡ | n/a | 4791 vs 6628 (15) | 7 | 0.80 (0.68 to 0.93) | .003 | 38 (0 to 74) | 0.86 | 7 | 0.98 0.79 to 1.22) | .97 | 0 (0 to 71) | 0.06 | 0.57 | ABB |
| ΝΑΤ2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | 15 | 1.01 (0.83 to 1.22) | .94 | 81 (70 to 88) | 0.06 | 15 | 0.95 (0.76 to 1.20) | .68 | 64 (37 to 79) | 0.17 | 0.98 | CCB |
| NQO1 | Pro187Ser(C609T) | rs1800566 | 5084 vs 5932 (8) | 8 | 1.14 (0.96 to 1.35) | .12 | 64 (22 to 83) | 0.89 | 8 | 1.10 (0.76 to 1.59) | .63 | 56 (2 to 80) | 0.19 | 0.95 | ACB |
| Tumor suppressor genes | |||||||||||||||
| TP53 | Arg72Pro|||| | rs1042522 | 7414 vs 9872 (27) | 27 | 1.01 (0.89 to 1.14) | .90 | — | — | 27 | 1.04 (0.82 to 1.31) | .77 | — | — | 0.99 | --C |
| TP53 | intron3 16bp|||| | rs17878362 | 1637 vs 1874 (5) | 5 | 1.58 (0.98 to 2.56) | — | — | — | 5 | 1.14 (0.84 to 1.55) | — | — | — | 0.92 | --C |
| MDM2 | 309 T/G|||| | rs2279744 | 2543 vs 2115 (7) | 7 | 0.73 (0.62 to 0.86) | — | — | — | 7 | 0.86 (0.57 to 1.30) | — | — | — | 0.10 | --C |
| Vitamin D and calcium metabolism | |||||||||||||||
| VDR | BsmI(60890GA) | rs1544410‡ | 5607 vs 6202 (7) | 7 | 0.77 (0.58 to 1.02) | .07 | 89 (81 to 94) | 1.00 | 7 | 0.51 (0.28 to 0.90) | .02 | 95 (92 to 97) | 1.00 | 0.92 | ACB |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9||) | 9 | 0.97 (0.85 to 1.11) | .66 | 67 (32 to 83) | 0.16 | 9 | 0.99 (0.82 to 1.20) | .94 | 70 (41 to 85) | 0.06 | 0.98 | CCB |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | 4 | 1.06 (0.87 to 1.30) | .55 | 0 (0 to 85) | 0.09 | 3 | 1.00 (0.57 to 1.75) | .99 | 73 (8 to 92) | 0.05 | 0.97 | CAB |
* BFDP value for the heterozygous additive model (var/wt VS wt/wt) at prior probability of 0.05. BFDP level of noteworthiness 0.2. For more information please see Supplementary Table 6
† Venice criteria grade for the the heterozygous additive model (var/wt VS wt/wt). For the third criterion (protection from bias), all meta-analyses of candidate gene association studies were scored with B. There was no obvious bias in the studies included, but there was considerable missing information on the generation of evidence. For the published meta-analyses, from which neither power nor I2 could be determined, we considered that there is considerable potential for bias (there were few small studies), so that third category would be “C.”
‡ The associations considered to be “less-credible positives”.
§ Includes unpublished data from Scottish GWAS.|| Includes unpublished data from Canadian GWAS.¶ (IV) Liu et al. 2011 (131).
# McColgan and Sharma 2009 (64).
** Zhong et al. 2010 (137).†† The associations considered to be “positives”.
‡‡ Fang et al. 2010 (76).
§§ Economopoulos and Sergentanis 2010 (92).
|||| Economopoulos et al. 2010 (119).
¶¶ Hu et al. 2010 (144).
## Fang et al. 2011 (145).
Summary crude odds ratios (ORs) and 95% confidence intervals (95% CIs) for two additive models for variants that were identified from candidate studies
| Gene . | Variant . | rs number . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | I2(95% CI) . | Power . | N . | OR (95% CI) . | P . | I2(95% CI) . | Power . | BFDP* . | Venice criteria grade† . | N . | ||||
| Adhesion molecules | |||||||||||||||
| CDH1 | C-160A‡ | rs16260 | 7493 vs 7329 (5§) | 5 | 0.91 (0.85 to 0.97) | .005 | 49 (0 to 81) | 0.78 | 5 | 0.84 (0.74 to 0.96) | .01 | 34 (0 to 75) | 0.76 | 0.67 | BBB |
| MMP1 | G-1607GG‡ | rs1799750 | 1007 vs 1032 (5) | 5 | 1.05 (0.84 to 1.33) | .66 | 15 (0 to 82) | 0.07 | 5 | 1.54 (1.00 to 2.37) | .05 | 53 (0 to 83) | 0.61 | 0.88 | BBB |
| MMP3 | AAAAA-612AAAAAA‡ | rs3025058 | 857 vs 932 (4) | 4 | 0.79 (0.60 to 1.03) | .08 | 0 (0 to 85) | 0.43 | 4 | 1.16 (0.86 to 1.56) | .33 | 0 (0 to 85) | 0.20 | 0.92 | CAB |
| MMP9 | 1562C/T|| | rs3918242 | 575 vs 836 (4) | — | — | — | — | — | — | — | — | — | — | — | — |
| Alcohol metabolism | |||||||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | 5 | 1.06 (0.83 to 1.36) | .63 | 70 (24 to 88) | 0.15 | 5 | 1.22 (0.94 to 1.58) | .13 | 26 (0 to 70) | 0.35 | 0.97 | CCB |
| ADH1C | Ile349Val(1045A>G) | rs698 | 3168 vs 6229 (7) | 7 | 0.96 (0.87 to 1.07) | .49 | 0 (0 to 78) | 0.12 | 7 | 0.88 (0.66 to 1.16) | .35 | 65 (21 to 84) | 0.4 | 0.98 | CBB |
| ALDH2 | Glu487Lys | rs671‡ | 2209 vs 3383 (8) | 8 | 0.88 (0.79 to 0.99) | .04 | 0 (0 to 65) | 0.59 | 8 | 0.89 (0.58 to 1.36) | .59 | 58 (12 to 80) | 0.17 | 0.89 | CCB |
| Angiogenesis | |||||||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | 4 | 0.94 (0.71 to 1.24) | .69 | 55 (0 to 85) | 0.13 | 4 | 1.19 (0.72 to 1.99) | .50 | 15 (0 to 87) | 0.10 | 0.97 | CBB |
| VEGF | G634C¶ | rs2010963 | 1508 vs 1308 (4) | 4 | 0.89 (0.72 to 1.10) | — | — | — | 4 | 1.17 (0.93 to 1.47) | — | — | — | 0.96 | --C |
| Base-excision repair | |||||||||||||||
| MGMT | Leu84Phe** | rs12917 | 1524 vs 4646 (5) | 5 | 0.84 (0.70 to 1.00) | .05 | — | — | 5 | 0.97 (0.56 to 1.66) | .97 | — | — | 0.91 | --C |
| MGMT | Il3143Val** | rs2308321 | 1326 vs 3520 (4) | 4 | 0.86 (0.66 to 1.12) | .26 | — | — | 4 | 1.01 (0.56 to 1.81) | .69 | — | — | 0.91 | --C |
| MUTYH | G396D | rs36053993†† | 26 592 vs 19 207 (15) | 15 | 1.07 (0.91 to 1.27) | .41 | 0 (0 to 54) | 0.13 | 9 | 6.15 (2.34 to 16.15) | .00 | 0 (0 to 65) | 0.50 | 0.17 | CAB |
| MUTYH | Y179C | rs34612342‡ | 26 370 vs 19 042 (15) | 14 | 1.34 (1.02 to 1.77) | .04 | 0 (0 to 55) | 0.55 | 6 | 3.35 (1.14 to 9.89) | .03 | 0 (0 to 75) | 0.37 | 0.89 | BAB |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | 9 | 1.02 (0.94 to 1.12) | .60 | 48 (0 to 76) | 0.08 | 9 | 1.05 (0.89 to 1.23) | .58 | 42 (0 to 73) | 0.14 | 0.99 | CBB |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11§) | 10 | 0.96 (0.87 to 1.07) | .50 | 0 (0 to 62) | 0.13 | 10 | 1.10 (0.82 to 1.48) | .52 | 16 (0 to 57) | 0.13 | 0.98 | CAB |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | 4 | 1.06 (0.83 to 1.34) | .65 | 41 (0 to 80) | 0.08 | 3 | 1.16 (0.37 to 3.62) | .31 | 14 (0 to 91) | 0.06 | 0.97 | CBB |
| XRCC1 | Arg399Gln | rs25487‡ | 7247 vs 8786 (12§) | 12 | 0.99 (0.92 to 1.06) | .72 | 11 (0 to 51) | 0.06 | 12 | 0.88 (0.79 to 0.97) | .02 | 0 (0 to 58) | 0.67 | 0.99 | BAB |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10§) | 10 | 0.92 (0.84 to 1.01) | .09 | 36 (0 to 69) | 0.48 | 10 | 0.95 (0.72 to 1.24) | .68 | 57 (12 to 79) | 0.12 | 0.95 | CBB |
| Inflammation or immune response | |||||||||||||||
| IL6 | 174G>C | rs1800795‡ | 6676 vs 7942 (10§,||) | 10 | 1.03 (0.91 to 1.17) | .65 | 56 (11 to 78) | 0.13 | 10 | 0.94 (0.79 to 1.12) | .48 | 56 (11 to 78) | 0.24 | 0.98 | CCB |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7§) | 7 | 1.03 (0.92 to 1.15) | .61 | 44 (0 to 77) | 0.08 | 7 | 1.05 (0.91 to 1.20) | .53 | 0 (0 to 71) | 0.11 | 0.99 | CBB |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5§) | 5 | 0.96 (0.85 to 1.08) | .49 | 0 (0 to 79) | 0.11 | 5 | 0.93 (0.81 to 1.07) | .30 | 35 (0 to 75) | 0.18 | 0.98 | CAB |
| PPARγ | C1431T | rs3856806 | 5574 vs 7035 (7§) | 5 | 1.04 (0.95 to 1.13) | .44 | 0 (0 to 79) | 0.14 | 5 | 0.95 (0.50 to 1.78) | .86 | 69 (22 to 88) | 0.06 | 0.98 | CAB |
| PPARγ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17§,||) | 13 | 0.98 (0.86 to 1.11) | .72 | 65 (36 to 80) | 0.09 | 12 | 0.91 (0.73 to 1.12) | .37 | 0 (0 to 58) | 0.14 | 0.98 | CCB |
| PTGS2 | A1195G | rs689466 | 4756 vs 6030 (7§) | 7 | 1.04 (0.95 to 1.13) | .42 | 41 (0 to 75) | 0.16 | 7 | 1.08 (0.77 to 1.51) | .66 | 64 (19 to 84) | 0.19 | 0.98 | CBB |
| PTGS2 | A1803G | rs4648298 | 4229 vs 4279 (5§) | 3 | 1.00 (0.83 to 1.22) | .96 | 49 (0 to 85) | 0.05 | 3 | n/a | n/a | n/a | n/a | 0.98 | CBB |
| PTGS2 | C427T | rs5275 | 4745 vs 5756 (7§) | 7 | 1.01 (0.93 to 1.09) | .87 | 0 (0 to 71) | 0.06 | 7 | 1.03 (0.91 to 1.17) | .65 | 0 (0 to 71) | 0.08 | 0.99 | CAB |
| PTGS2 | G306C | rs5277 | 4269 vs 4735 (5§) | 5 | 0.97 (0.88 to 1.06) | .45 | 23 (0 to 68) | 0.10 | 5 | 0.85 (0.65 to 1.11) | .24 | 5 (0 to 80) | 0.23 | 0.99 | CAB |
| PTGS2 | G765C | rs20417 | 5459 vs 7272 (11§) | 10 | 1.03 (0.95 to 1.13) | .45 | 45 (0 to 74) | 0.11 | 8 | 1.21 (0.93 to 1.57) | .15 | 0 (0 to 68) | 0.3 | 0.99 | CBB |
| PTGS2 | T1532C | rs5273 | 2843 vs 3216 (5§) | 2 | n/a | n/a | n/a | n/a | 2 | n/a | n/a | n/a | n/a | — | — |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9§) | 9 | 1.11 (0.88 to 1.40) | .37 | 72 (45 to 86) | 0.65 | 9 | 1.13 (0.92 to 1.39) | .24 | 47 (0 to 75) | 0.21 | 0.97 | BCB |
| NOD2 | 3020incC‡ | rs5743293 | 4222 vs 2988 (8) | 8 | 1.39 (1.15 to 1.69) | .001 | 0 (0 to 68) | 0.94 | 5 | 2.81 (0.87 to 9.05) | .08 | 0 (0 to 79) | 0.26 | 0.34 | AAB |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6§) | 6 | 1.41 (1.04 to 1.91) | .03 | 0 (0 to 75) | 0.63 | 0 | n/a | n/a | n/a | n/a | 0.87 | BBB |
| NOD2 | R702W‡ | rs2066844 | 3445 vs 2731 (6§) | 6 | 1.22 (1.00 to 1.50) | .06 | 12 (0 to 78) | 0.49 | 4 | 1.23 (0.41 to 3.70) | .71 | 0 (0 to 85) | 0.06 | 0.91 | BAB |
| Inhibition of cell growth | |||||||||||||||
| CCND1 | 870A‡ | rs17852153 | 4747 vs 6783 (13) | 13 | 1.13 (1.03 to 1.25) | .02 | 0 (0 to 57) | 0.70 | 13 | 1.16 (0.98 to 1.38) | .09 | 51 (8 to 74) | 0.78 | 0.85 | BAB |
| TGFB1 | C509T‡‡ | rs1800469 | 994 vs 2335 (5) | 5 | 1.12 (0.91 to 1.37) | — | — | — | 5 | 1.62 (1.30 to 2.02) | — | — | — | 0.96 | --C |
| TGFBR1 | TGFBR1*6A‡ | rs11466445 | 3217 vs 4539 (8) | 8 | 1.15 (1.01 to 1.31) | .03 | 0 (0 to 68) | — | 8 | 1.51 (0.69 to 3.31) | .31 | 52 (0 to 87) | — | 0.89 | -AB |
| Insulin-related | |||||||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 6 | 1.06 (0.99 to 1.14) | .08 | 0 (0 to 75) | 0.4 | 6 | 1.07 (0.97 to 1.18) | .15 | 0 (0 to 75) | 0.31 | 0.97 | CAB |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | 6 | 1.02 (0.94 to 1.10) | .72 | 0 (0 to 75) | 0.08 | 6 | 1.00 (0.91 to 1.10) | .96 | 0 (0 to 75) | 0.05 | 0.99 | CAB |
| Iron metabolism | |||||||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6§,||) | 6 | 1.08 (0.97 to 1.21) | .15 | 0 (0 to 75) | 0.29 | 5 | 0.90 (0.55 to 1.48) | .69 | 0 (0 to 79) | 0.07 | 0.97 | CAB |
| Lipid metabolism | |||||||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5§) | 5 | 0.94 (0.85 to 1.04) | .23 | 66 (11 to 87) | 0.21 | 4 | 0.75 (0.46 to 1.22) | .25 | 0 (0 to 85) | 0.21 | 0.98 | CCB |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5§) | 5 | 1.04 (0.94 to 1.15) | .49 | 14 (0 to 82) | 0.12 | 5 | 1.12 (0.83 to 1.52) | .45 | 0 (0 to 79) | 0.12 | 0.98 | CAB |
| Mitotic control | |||||||||||||||
| STK15 | F31I | rs2273535 | 4860 vs 4629 (4) | 4 | 1.03 (0.94 to 1.12) | .58 | 0 (0 to 85) | 0.1 | 4 | 1.30 (0.96 to 1.76) | .09 | 52 (0 to 84) | 0.56 | 0.99 | CAB |
| One-carbon metabolism | |||||||||||||||
| MTHFR | C677T | rs1801133†† | 27 372 vs 39 867 (52||) | 52 | 1.00 (0.94 to 1.06) | .92 | 53 (36 to 65) | 0.05 | 52 | 0.87 (0.82 to 0.91) | .00 | 35 (10 to 53) | 1.00 | 0.99 | CBB |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34||) | 34 | 1.01 (0.97 to 1.06) | .51 | 0 (0 to 37) | 0.08 | 34 | 0.94 (0.87 to 1.01) | .09 | 22 (0 to 49) | 0.40 | 0.99 | CAB |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14||) | 14 | 0.97 (0.92 to 1.02) | .27 | 12 (0 to 50) | 0.21 | 14 | 0.96 (0.84 to 1.09) | .50 | 48 (4 to 72) | 0.05 | 0.99 | CAB |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | 9 | 0.98 (0.90 to 1.07) | .66 | 15 (0 to 57) | 0.10 | 9 | 1.04 (0.94 to 1.14) | .47 | 23 (0 to 64) | 0.17 | 0.99 | CAB |
| TS | TSER‡ | rs34743033 | 3519 vs 5289 (5) | 5 | 0.86 (0.78 to 0.95) | .003 | 18 (0 to 83) | 0.87 | 5 | 0.83 (0.73 to 0.94) | .004 | 17 (0 to 83) | 0.85 | 0.55 | AAB |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | 4 | 0.96 (0.88 to 1.06) | .45 | 0 (0 to 85) | 0.13 | 4 | 1.03 (0.88 to 1.19) | .73 | 0 (0 to 85) | 0.07 | 0.98 | CAB |
| Rare, high penetrance | |||||||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | 6 | 1.13 (0.88 to 1.47) | .34 | 0 (0 to 75) | 0.15 | 1 | n/a | n/a | n/a | n/a | 0.96 | CAB |
| APC | D1822V‡ | rs459552 | 6282 vs 7038 (6) | 6 | 0.99 (0.92 to 1.07) | .83 | 24 (0 to 68) | 0.06 | 6 | 0.84 (0.71 to 0.98) | .03 | 0 (0 to 75) | 0.55 | 0.99 | CAB |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7§) | 7 | 1.09 (0.90 to 1.32) | .40 | 55 (0 to 81) | 0.42 | 7 | 1.01 (0.85 to 1.21) | .88 | 43 (0 to 76) | 0.05 | 0.97 | CCB |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6§) | 5 | 1.06 (0.97 to 1.15) | .23 | 0 (0 to 79) | 0.27 | 5 | 1.15 (0.95 to 1.39) | .15 | 26 (0 to 71) | 0.33 | 0.97 | CAB |
| Substrate metabolism | |||||||||||||||
| CYP1A1 | 2454A>G‡ | rs1048943 | 10 274 vs 11 978 (13§,||) | 13 | 1.28 (1.01 to 1.63) | .05 | 85 (7 to 91) | 1.00 | 12 | 1.47 (1.17 to 1.85) | .001 | 3 (0 to 60) | 0.93 | 0.89 | ACB |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | 7 | 0.94 (0.86 to 1.04) | .23 | 0 (0 to 71) | 0.27 | 7 | 0.84 (0.56 to 1.27) | .42 | 54 (0 to 80) | 0.38 | 0.98 | CAB |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | 9 | 1.13 (0.95 to 1.34) | .18 | 63 (25 to 82) | 0.75 | 9 | 1.07 (0.92 to 1.26) | .40 | 42 (0 to 73) | 0.16 | 0.96 | ACB |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6§) | 6 | 0.99 (0.92 to 1.06) | .69 | 0 (0 to 75) | 0.06 | 6 | 0.98 (0.90 to 1.07) | .70 | 0 (0 to 77) | 0.08 | 0.99 | CAB |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6§) | 6 | 0.93 (0.85 to 1.02) | .13 | 21 (0 to 65) | 0.34 | 6 | 1.29 (0.99 to 1.70) | .06 | 0 (0 to 75) | 0.46 | 0.97 | CAB |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6§) | 6 | 1.08 (0.83 to 1.40) | .57 | 73 (33 to 88) | 0.26 | 4 | 0.68 (0.37 to 1.26) | .22 | 11 (0 to 86) | 0.23 | 0.97 | CCB |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8||) | 8 | 0.93 (0.83 to 1.05) | .23 | 0 (0 to 68) | 0.27 | 8 | 1.23 (0.92 to 1.63) | .16 | 35 (0 to 71) | 0.34 | 0.97 | CAB |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | 7 | 1.17 (0.92 to 1.48) | .21 | 53 (0 to 80) | 0.61 | 6 | 1.83 (0.94 to 3.57) | .08 | 0 (0 to 75) | 0.55 | 0.95 | BCB |
| GSTA1 | GSTA1*B§§ | 1648 vs 2039 (4) | 4 | 1.03 (0.89 to 1.19) | — | — | — | 4 | 1.09 (0.90 to 1.32) | — | — | — | 0.98 | — | |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22§) | 22 | 1.05 (0.99 to 1.12) | .11 | 0 (0 to 46) | 0.38 | 22 | 0.95 (0.86 to 1.05) | .32 | 36 (0 to 62) | 0.17 | 0.98 | CAB |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6§,||) | 6 | 1.02 (0.91 to 1.13) | .77 | 0 (0 to 75) | 0.07 | 6 | 0.87 (0.55 to 1.37) | .55 | 12 (0 to 78) | 0.09 | 0.99 | CAB |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| ΝΑΤ1 | slow/rapid‡ | n/a | 4791 vs 6628 (15) | 7 | 0.80 (0.68 to 0.93) | .003 | 38 (0 to 74) | 0.86 | 7 | 0.98 0.79 to 1.22) | .97 | 0 (0 to 71) | 0.06 | 0.57 | ABB |
| ΝΑΤ2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | 15 | 1.01 (0.83 to 1.22) | .94 | 81 (70 to 88) | 0.06 | 15 | 0.95 (0.76 to 1.20) | .68 | 64 (37 to 79) | 0.17 | 0.98 | CCB |
| NQO1 | Pro187Ser(C609T) | rs1800566 | 5084 vs 5932 (8) | 8 | 1.14 (0.96 to 1.35) | .12 | 64 (22 to 83) | 0.89 | 8 | 1.10 (0.76 to 1.59) | .63 | 56 (2 to 80) | 0.19 | 0.95 | ACB |
| Tumor suppressor genes | |||||||||||||||
| TP53 | Arg72Pro|||| | rs1042522 | 7414 vs 9872 (27) | 27 | 1.01 (0.89 to 1.14) | .90 | — | — | 27 | 1.04 (0.82 to 1.31) | .77 | — | — | 0.99 | --C |
| TP53 | intron3 16bp|||| | rs17878362 | 1637 vs 1874 (5) | 5 | 1.58 (0.98 to 2.56) | — | — | — | 5 | 1.14 (0.84 to 1.55) | — | — | — | 0.92 | --C |
| MDM2 | 309 T/G|||| | rs2279744 | 2543 vs 2115 (7) | 7 | 0.73 (0.62 to 0.86) | — | — | — | 7 | 0.86 (0.57 to 1.30) | — | — | — | 0.10 | --C |
| Vitamin D and calcium metabolism | |||||||||||||||
| VDR | BsmI(60890GA) | rs1544410‡ | 5607 vs 6202 (7) | 7 | 0.77 (0.58 to 1.02) | .07 | 89 (81 to 94) | 1.00 | 7 | 0.51 (0.28 to 0.90) | .02 | 95 (92 to 97) | 1.00 | 0.92 | ACB |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9||) | 9 | 0.97 (0.85 to 1.11) | .66 | 67 (32 to 83) | 0.16 | 9 | 0.99 (0.82 to 1.20) | .94 | 70 (41 to 85) | 0.06 | 0.98 | CCB |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | 4 | 1.06 (0.87 to 1.30) | .55 | 0 (0 to 85) | 0.09 | 3 | 1.00 (0.57 to 1.75) | .99 | 73 (8 to 92) | 0.05 | 0.97 | CAB |
| Gene . | Variant . | rs number . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) . | P . | I2(95% CI) . | Power . | N . | OR (95% CI) . | P . | I2(95% CI) . | Power . | BFDP* . | Venice criteria grade† . | N . | ||||
| Adhesion molecules | |||||||||||||||
| CDH1 | C-160A‡ | rs16260 | 7493 vs 7329 (5§) | 5 | 0.91 (0.85 to 0.97) | .005 | 49 (0 to 81) | 0.78 | 5 | 0.84 (0.74 to 0.96) | .01 | 34 (0 to 75) | 0.76 | 0.67 | BBB |
| MMP1 | G-1607GG‡ | rs1799750 | 1007 vs 1032 (5) | 5 | 1.05 (0.84 to 1.33) | .66 | 15 (0 to 82) | 0.07 | 5 | 1.54 (1.00 to 2.37) | .05 | 53 (0 to 83) | 0.61 | 0.88 | BBB |
| MMP3 | AAAAA-612AAAAAA‡ | rs3025058 | 857 vs 932 (4) | 4 | 0.79 (0.60 to 1.03) | .08 | 0 (0 to 85) | 0.43 | 4 | 1.16 (0.86 to 1.56) | .33 | 0 (0 to 85) | 0.20 | 0.92 | CAB |
| MMP9 | 1562C/T|| | rs3918242 | 575 vs 836 (4) | — | — | — | — | — | — | — | — | — | — | — | — |
| Alcohol metabolism | |||||||||||||||
| ADH1B | Arg47His | rs1229984 | 1931 vs 2898 (5) | 5 | 1.06 (0.83 to 1.36) | .63 | 70 (24 to 88) | 0.15 | 5 | 1.22 (0.94 to 1.58) | .13 | 26 (0 to 70) | 0.35 | 0.97 | CCB |
| ADH1C | Ile349Val(1045A>G) | rs698 | 3168 vs 6229 (7) | 7 | 0.96 (0.87 to 1.07) | .49 | 0 (0 to 78) | 0.12 | 7 | 0.88 (0.66 to 1.16) | .35 | 65 (21 to 84) | 0.4 | 0.98 | CBB |
| ALDH2 | Glu487Lys | rs671‡ | 2209 vs 3383 (8) | 8 | 0.88 (0.79 to 0.99) | .04 | 0 (0 to 65) | 0.59 | 8 | 0.89 (0.58 to 1.36) | .59 | 58 (12 to 80) | 0.17 | 0.89 | CCB |
| Angiogenesis | |||||||||||||||
| VEGF | 936 C>T | rs3025039 | 1317 vs 1192 (4) | 4 | 0.94 (0.71 to 1.24) | .69 | 55 (0 to 85) | 0.13 | 4 | 1.19 (0.72 to 1.99) | .50 | 15 (0 to 87) | 0.10 | 0.97 | CBB |
| VEGF | G634C¶ | rs2010963 | 1508 vs 1308 (4) | 4 | 0.89 (0.72 to 1.10) | — | — | — | 4 | 1.17 (0.93 to 1.47) | — | — | — | 0.96 | --C |
| Base-excision repair | |||||||||||||||
| MGMT | Leu84Phe** | rs12917 | 1524 vs 4646 (5) | 5 | 0.84 (0.70 to 1.00) | .05 | — | — | 5 | 0.97 (0.56 to 1.66) | .97 | — | — | 0.91 | --C |
| MGMT | Il3143Val** | rs2308321 | 1326 vs 3520 (4) | 4 | 0.86 (0.66 to 1.12) | .26 | — | — | 4 | 1.01 (0.56 to 1.81) | .69 | — | — | 0.91 | --C |
| MUTYH | G396D | rs36053993†† | 26 592 vs 19 207 (15) | 15 | 1.07 (0.91 to 1.27) | .41 | 0 (0 to 54) | 0.13 | 9 | 6.15 (2.34 to 16.15) | .00 | 0 (0 to 65) | 0.50 | 0.17 | CAB |
| MUTYH | Y179C | rs34612342‡ | 26 370 vs 19 042 (15) | 14 | 1.34 (1.02 to 1.77) | .04 | 0 (0 to 55) | 0.55 | 6 | 3.35 (1.14 to 9.89) | .03 | 0 (0 to 75) | 0.37 | 0.89 | BAB |
| OGG1 | Ser326Cys | rs1052133 | 4713 vs 6165 (9) | 9 | 1.02 (0.94 to 1.12) | .60 | 48 (0 to 76) | 0.08 | 9 | 1.05 (0.89 to 1.23) | .58 | 42 (0 to 73) | 0.14 | 0.99 | CBB |
| XRCC1 | Arg194Trp | rs1799782 | 6635 vs 8488 (11§) | 10 | 0.96 (0.87 to 1.07) | .50 | 0 (0 to 62) | 0.13 | 10 | 1.10 (0.82 to 1.48) | .52 | 16 (0 to 57) | 0.13 | 0.98 | CAB |
| XRCC1 | Arg280His | rs25489 | 3114 vs 3679 (5) | 4 | 1.06 (0.83 to 1.34) | .65 | 41 (0 to 80) | 0.08 | 3 | 1.16 (0.37 to 3.62) | .31 | 14 (0 to 91) | 0.06 | 0.97 | CBB |
| XRCC1 | Arg399Gln | rs25487‡ | 7247 vs 8786 (12§) | 12 | 0.99 (0.92 to 1.06) | .72 | 11 (0 to 51) | 0.06 | 12 | 0.88 (0.79 to 0.97) | .02 | 0 (0 to 58) | 0.67 | 0.99 | BAB |
| XRCC3 | Thr241Met | rs861539 | 4484 vs 5235 (10§) | 10 | 0.92 (0.84 to 1.01) | .09 | 36 (0 to 69) | 0.48 | 10 | 0.95 (0.72 to 1.24) | .68 | 57 (12 to 79) | 0.12 | 0.95 | CBB |
| Inflammation or immune response | |||||||||||||||
| IL6 | 174G>C | rs1800795‡ | 6676 vs 7942 (10§,||) | 10 | 1.03 (0.91 to 1.17) | .65 | 56 (11 to 78) | 0.13 | 10 | 0.94 (0.79 to 1.12) | .48 | 56 (11 to 78) | 0.24 | 0.98 | CCB |
| IL8 | 251T/A | rs4073 | 3228 vs 3772 (7§) | 7 | 1.03 (0.92 to 1.15) | .61 | 44 (0 to 77) | 0.08 | 7 | 1.05 (0.91 to 1.20) | .53 | 0 (0 to 71) | 0.11 | 0.99 | CBB |
| IL10 | 1082G/A | rs1800896 | 2964 vs 3621 (5§) | 5 | 0.96 (0.85 to 1.08) | .49 | 0 (0 to 79) | 0.11 | 5 | 0.93 (0.81 to 1.07) | .30 | 35 (0 to 75) | 0.18 | 0.98 | CAB |
| PPARγ | C1431T | rs3856806 | 5574 vs 7035 (7§) | 5 | 1.04 (0.95 to 1.13) | .44 | 0 (0 to 79) | 0.14 | 5 | 0.95 (0.50 to 1.78) | .86 | 69 (22 to 88) | 0.06 | 0.98 | CAB |
| PPARγ | Pro12Ala | rs1801282 | 15 091 vs 18 690 (17§,||) | 13 | 0.98 (0.86 to 1.11) | .72 | 65 (36 to 80) | 0.09 | 12 | 0.91 (0.73 to 1.12) | .37 | 0 (0 to 58) | 0.14 | 0.98 | CCB |
| PTGS2 | A1195G | rs689466 | 4756 vs 6030 (7§) | 7 | 1.04 (0.95 to 1.13) | .42 | 41 (0 to 75) | 0.16 | 7 | 1.08 (0.77 to 1.51) | .66 | 64 (19 to 84) | 0.19 | 0.98 | CBB |
| PTGS2 | A1803G | rs4648298 | 4229 vs 4279 (5§) | 3 | 1.00 (0.83 to 1.22) | .96 | 49 (0 to 85) | 0.05 | 3 | n/a | n/a | n/a | n/a | 0.98 | CBB |
| PTGS2 | C427T | rs5275 | 4745 vs 5756 (7§) | 7 | 1.01 (0.93 to 1.09) | .87 | 0 (0 to 71) | 0.06 | 7 | 1.03 (0.91 to 1.17) | .65 | 0 (0 to 71) | 0.08 | 0.99 | CAB |
| PTGS2 | G306C | rs5277 | 4269 vs 4735 (5§) | 5 | 0.97 (0.88 to 1.06) | .45 | 23 (0 to 68) | 0.10 | 5 | 0.85 (0.65 to 1.11) | .24 | 5 (0 to 80) | 0.23 | 0.99 | CAB |
| PTGS2 | G765C | rs20417 | 5459 vs 7272 (11§) | 10 | 1.03 (0.95 to 1.13) | .45 | 45 (0 to 74) | 0.11 | 8 | 1.21 (0.93 to 1.57) | .15 | 0 (0 to 68) | 0.3 | 0.99 | CBB |
| PTGS2 | T1532C | rs5273 | 2843 vs 3216 (5§) | 2 | n/a | n/a | n/a | n/a | 2 | n/a | n/a | n/a | n/a | — | — |
| TNF-α | 308G>A | rs1800629 | 3843 vs 4098 (9§) | 9 | 1.11 (0.88 to 1.40) | .37 | 72 (45 to 86) | 0.65 | 9 | 1.13 (0.92 to 1.39) | .24 | 47 (0 to 75) | 0.21 | 0.97 | BCB |
| NOD2 | 3020incC‡ | rs5743293 | 4222 vs 2988 (8) | 8 | 1.39 (1.15 to 1.69) | .001 | 0 (0 to 68) | 0.94 | 5 | 2.81 (0.87 to 9.05) | .08 | 0 (0 to 79) | 0.26 | 0.34 | AAB |
| NOD2 | G908R | rs2066845 | 4541 vs 3820 (6§) | 6 | 1.41 (1.04 to 1.91) | .03 | 0 (0 to 75) | 0.63 | 0 | n/a | n/a | n/a | n/a | 0.87 | BBB |
| NOD2 | R702W‡ | rs2066844 | 3445 vs 2731 (6§) | 6 | 1.22 (1.00 to 1.50) | .06 | 12 (0 to 78) | 0.49 | 4 | 1.23 (0.41 to 3.70) | .71 | 0 (0 to 85) | 0.06 | 0.91 | BAB |
| Inhibition of cell growth | |||||||||||||||
| CCND1 | 870A‡ | rs17852153 | 4747 vs 6783 (13) | 13 | 1.13 (1.03 to 1.25) | .02 | 0 (0 to 57) | 0.70 | 13 | 1.16 (0.98 to 1.38) | .09 | 51 (8 to 74) | 0.78 | 0.85 | BAB |
| TGFB1 | C509T‡‡ | rs1800469 | 994 vs 2335 (5) | 5 | 1.12 (0.91 to 1.37) | — | — | — | 5 | 1.62 (1.30 to 2.02) | — | — | — | 0.96 | --C |
| TGFBR1 | TGFBR1*6A‡ | rs11466445 | 3217 vs 4539 (8) | 8 | 1.15 (1.01 to 1.31) | .03 | 0 (0 to 68) | — | 8 | 1.51 (0.69 to 3.31) | .31 | 52 (0 to 87) | — | 0.89 | -AB |
| Insulin-related | |||||||||||||||
| IGF1 | CA-repeat | n/a | 7900 vs 9161 (6) | 6 | 1.06 (0.99 to 1.14) | .08 | 0 (0 to 75) | 0.4 | 6 | 1.07 (0.97 to 1.18) | .15 | 0 (0 to 75) | 0.31 | 0.97 | CAB |
| IGFBP3 | 202A>C | rs2854744 | 7296 vs 10 452 (6) | 6 | 1.02 (0.94 to 1.10) | .72 | 0 (0 to 75) | 0.08 | 6 | 1.00 (0.91 to 1.10) | .96 | 0 (0 to 75) | 0.05 | 0.99 | CAB |
| Iron metabolism | |||||||||||||||
| HFE | C282T | rs1800562 | 5177 vs 6150 (6§,||) | 6 | 1.08 (0.97 to 1.21) | .15 | 0 (0 to 75) | 0.29 | 5 | 0.90 (0.55 to 1.48) | .69 | 0 (0 to 79) | 0.07 | 0.97 | CAB |
| Lipid metabolism | |||||||||||||||
| ApoE | e2 | rs7412 | 5821 vs 6754 (5§) | 5 | 0.94 (0.85 to 1.04) | .23 | 66 (11 to 87) | 0.21 | 4 | 0.75 (0.46 to 1.22) | .25 | 0 (0 to 85) | 0.21 | 0.98 | CCB |
| ApoE | e4 | rs429358 | 3808 vs 4684 (5§) | 5 | 1.04 (0.94 to 1.15) | .49 | 14 (0 to 82) | 0.12 | 5 | 1.12 (0.83 to 1.52) | .45 | 0 (0 to 79) | 0.12 | 0.98 | CAB |
| Mitotic control | |||||||||||||||
| STK15 | F31I | rs2273535 | 4860 vs 4629 (4) | 4 | 1.03 (0.94 to 1.12) | .58 | 0 (0 to 85) | 0.1 | 4 | 1.30 (0.96 to 1.76) | .09 | 52 (0 to 84) | 0.56 | 0.99 | CAB |
| One-carbon metabolism | |||||||||||||||
| MTHFR | C677T | rs1801133†† | 27 372 vs 39 867 (52||) | 52 | 1.00 (0.94 to 1.06) | .92 | 53 (36 to 65) | 0.05 | 52 | 0.87 (0.82 to 0.91) | .00 | 35 (10 to 53) | 1.00 | 0.99 | CBB |
| MTHFR | A1298C | rs1801131 | 17 178 vs 24 792 (34||) | 34 | 1.01 (0.97 to 1.06) | .51 | 0 (0 to 37) | 0.08 | 34 | 0.94 (0.87 to 1.01) | .09 | 22 (0 to 49) | 0.40 | 0.99 | CAB |
| MTR | A2756G | rs1805087 | 11 829 vs 15 975 (14||) | 14 | 0.97 (0.92 to 1.02) | .27 | 12 (0 to 50) | 0.21 | 14 | 0.96 (0.84 to 1.09) | .50 | 48 (4 to 72) | 0.05 | 0.99 | CAB |
| MTRR | A66G | rs1801394 | 6170 vs 8732 (9) | 9 | 0.98 (0.90 to 1.07) | .66 | 15 (0 to 57) | 0.10 | 9 | 1.04 (0.94 to 1.14) | .47 | 23 (0 to 64) | 0.17 | 0.99 | CAB |
| TS | TSER‡ | rs34743033 | 3519 vs 5289 (5) | 5 | 0.86 (0.78 to 0.95) | .003 | 18 (0 to 83) | 0.87 | 5 | 0.83 (0.73 to 0.94) | .004 | 17 (0 to 83) | 0.85 | 0.55 | AAB |
| TS | Ts1494del6 | rs34489327 | 3262 vs 4518 (4) | 4 | 0.96 (0.88 to 1.06) | .45 | 0 (0 to 85) | 0.13 | 4 | 1.03 (0.88 to 1.19) | .73 | 0 (0 to 85) | 0.07 | 0.98 | CAB |
| Rare, high penetrance | |||||||||||||||
| APC | E1317Q | rs1801166 | 6898 vs 6668 (6) | 6 | 1.13 (0.88 to 1.47) | .34 | 0 (0 to 75) | 0.15 | 1 | n/a | n/a | n/a | n/a | 0.96 | CAB |
| APC | D1822V‡ | rs459552 | 6282 vs 7038 (6) | 6 | 0.99 (0.92 to 1.07) | .83 | 24 (0 to 68) | 0.06 | 6 | 0.84 (0.71 to 0.98) | .03 | 0 (0 to 75) | 0.55 | 0.99 | CAB |
| MLH1 | I219V | rs1799977 | 2956 vs 5071 (7§) | 7 | 1.09 (0.90 to 1.32) | .40 | 55 (0 to 81) | 0.42 | 7 | 1.01 (0.85 to 1.21) | .88 | 43 (0 to 76) | 0.05 | 0.97 | CCB |
| MLH1 | -93 G>A | rs1800734 | 4524 vs 5544 (6§) | 5 | 1.06 (0.97 to 1.15) | .23 | 0 (0 to 79) | 0.27 | 5 | 1.15 (0.95 to 1.39) | .15 | 26 (0 to 71) | 0.33 | 0.97 | CAB |
| Substrate metabolism | |||||||||||||||
| CYP1A1 | 2454A>G‡ | rs1048943 | 10 274 vs 11 978 (13§,||) | 13 | 1.28 (1.01 to 1.63) | .05 | 85 (7 to 91) | 1.00 | 12 | 1.47 (1.17 to 1.85) | .001 | 3 (0 to 60) | 0.93 | 0.89 | ACB |
| CYP1A1 | 3698T>C | rs4646903 | 4897 vs 6559 (7) | 7 | 0.94 (0.86 to 1.04) | .23 | 0 (0 to 71) | 0.27 | 7 | 0.84 (0.56 to 1.27) | .42 | 54 (0 to 80) | 0.38 | 0.98 | CAB |
| CYP1A2 | 163C>A | rs762551 | 3051 vs 5326 (9) | 9 | 1.13 (0.95 to 1.34) | .18 | 63 (25 to 82) | 0.75 | 9 | 1.07 (0.92 to 1.26) | .40 | 42 (0 to 73) | 0.16 | 0.96 | ACB |
| CYP1B1 | 4326C>G | rs1056836 | 8514 vs 9721 (6§) | 6 | 0.99 (0.92 to 1.06) | .69 | 0 (0 to 75) | 0.06 | 6 | 0.98 (0.90 to 1.07) | .70 | 0 (0 to 77) | 0.08 | 0.99 | CAB |
| CYP2C9 | 430C>T | rs1799853 | 5134 vs 6164 (6§) | 6 | 0.93 (0.85 to 1.02) | .13 | 21 (0 to 65) | 0.34 | 6 | 1.29 (0.99 to 1.70) | .06 | 0 (0 to 75) | 0.46 | 0.97 | CAB |
| CYP2C9 | 1057A>C | rs1057910 | 5379 vs 6531 (6§) | 6 | 1.08 (0.83 to 1.40) | .57 | 73 (33 to 88) | 0.26 | 4 | 0.68 (0.37 to 1.26) | .22 | 11 (0 to 86) | 0.23 | 0.97 | CCB |
| CYP2E1 | 1053C>T | rs2031920 | 4456 vs 5077 (8||) | 8 | 0.93 (0.83 to 1.05) | .23 | 0 (0 to 68) | 0.27 | 8 | 1.23 (0.92 to 1.63) | .16 | 35 (0 to 71) | 0.34 | 0.97 | CAB |
| CYP2E1 | 1293G>C | rs3813867 | 3424 vs 4686 (7) | 7 | 1.17 (0.92 to 1.48) | .21 | 53 (0 to 80) | 0.61 | 6 | 1.83 (0.94 to 3.57) | .08 | 0 (0 to 75) | 0.55 | 0.95 | BCB |
| GSTA1 | GSTA1*B§§ | 1648 vs 2039 (4) | 4 | 1.03 (0.89 to 1.19) | — | — | — | 4 | 1.09 (0.90 to 1.32) | — | — | — | 0.98 | — | |
| GSTM1 | Null variant | n/a | 18 845 vs 26 662 (43) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| GSTP1 | IIe105Val | rs1695 | 9267 vs 12 902 (22§) | 22 | 1.05 (0.99 to 1.12) | .11 | 0 (0 to 46) | 0.38 | 22 | 0.95 (0.86 to 1.05) | .32 | 36 (0 to 62) | 0.17 | 0.98 | CAB |
| GSTP1 | Ala114Val | rs1138272 | 5183 vs 5457 (6§,||) | 6 | 1.02 (0.91 to 1.13) | .77 | 0 (0 to 75) | 0.07 | 6 | 0.87 (0.55 to 1.37) | .55 | 12 (0 to 78) | 0.09 | 0.99 | CAB |
| GSTT1 | Null variant | n/a | 13 410 vs 20 455 (35) | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | — |
| ΝΑΤ1 | slow/rapid‡ | n/a | 4791 vs 6628 (15) | 7 | 0.80 (0.68 to 0.93) | .003 | 38 (0 to 74) | 0.86 | 7 | 0.98 0.79 to 1.22) | .97 | 0 (0 to 71) | 0.06 | 0.57 | ABB |
| ΝΑΤ2 | slow/rapid | n/a | 12 908 vs 16 483 (26) | 15 | 1.01 (0.83 to 1.22) | .94 | 81 (70 to 88) | 0.06 | 15 | 0.95 (0.76 to 1.20) | .68 | 64 (37 to 79) | 0.17 | 0.98 | CCB |
| NQO1 | Pro187Ser(C609T) | rs1800566 | 5084 vs 5932 (8) | 8 | 1.14 (0.96 to 1.35) | .12 | 64 (22 to 83) | 0.89 | 8 | 1.10 (0.76 to 1.59) | .63 | 56 (2 to 80) | 0.19 | 0.95 | ACB |
| Tumor suppressor genes | |||||||||||||||
| TP53 | Arg72Pro|||| | rs1042522 | 7414 vs 9872 (27) | 27 | 1.01 (0.89 to 1.14) | .90 | — | — | 27 | 1.04 (0.82 to 1.31) | .77 | — | — | 0.99 | --C |
| TP53 | intron3 16bp|||| | rs17878362 | 1637 vs 1874 (5) | 5 | 1.58 (0.98 to 2.56) | — | — | — | 5 | 1.14 (0.84 to 1.55) | — | — | — | 0.92 | --C |
| MDM2 | 309 T/G|||| | rs2279744 | 2543 vs 2115 (7) | 7 | 0.73 (0.62 to 0.86) | — | — | — | 7 | 0.86 (0.57 to 1.30) | — | — | — | 0.10 | --C |
| Vitamin D and calcium metabolism | |||||||||||||||
| VDR | BsmI(60890GA) | rs1544410‡ | 5607 vs 6202 (7) | 7 | 0.77 (0.58 to 1.02) | .07 | 89 (81 to 94) | 1.00 | 7 | 0.51 (0.28 to 0.90) | .02 | 95 (92 to 97) | 1.00 | 0.92 | ACB |
| VDR | FokI | rs10735810 | 7646 vs 8968 (9||) | 9 | 0.97 (0.85 to 1.11) | .66 | 67 (32 to 83) | 0.16 | 9 | 0.99 (0.82 to 1.20) | .94 | 70 (41 to 85) | 0.06 | 0.98 | CCB |
| VDR | TaqI | rs731236 | 946 vs 1184 (4) | 4 | 1.06 (0.87 to 1.30) | .55 | 0 (0 to 85) | 0.09 | 3 | 1.00 (0.57 to 1.75) | .99 | 73 (8 to 92) | 0.05 | 0.97 | CAB |
* BFDP value for the heterozygous additive model (var/wt VS wt/wt) at prior probability of 0.05. BFDP level of noteworthiness 0.2. For more information please see Supplementary Table 6
† Venice criteria grade for the the heterozygous additive model (var/wt VS wt/wt). For the third criterion (protection from bias), all meta-analyses of candidate gene association studies were scored with B. There was no obvious bias in the studies included, but there was considerable missing information on the generation of evidence. For the published meta-analyses, from which neither power nor I2 could be determined, we considered that there is considerable potential for bias (there were few small studies), so that third category would be “C.”
‡ The associations considered to be “less-credible positives”.
§ Includes unpublished data from Scottish GWAS.|| Includes unpublished data from Canadian GWAS.¶ (IV) Liu et al. 2011 (131).
# McColgan and Sharma 2009 (64).
** Zhong et al. 2010 (137).†† The associations considered to be “positives”.
‡‡ Fang et al. 2010 (76).
§§ Economopoulos and Sergentanis 2010 (92).
|||| Economopoulos et al. 2010 (119).
¶¶ Hu et al. 2010 (144).
## Fang et al. 2011 (145).
Summary crude odds ratios (ORs) and 95% confidence intervals (95% CIs) for two additive models for variants that were identified from genome-wide association studies
| Gene . | Variant . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | BFDP* . | Venice criteria grade† . | |||
| Common, low penetrance | ||||||||||||||
| SMAD7‡ | rs4939827 | 37 650 vs 36 154 (13§) | 13 | 0.89 (0.86 to 0.92) | 3.7 × 10-12 | 0 (0 to 57) | 1.00 | 13 | 0.75 (0.71 to 0.79) | 3.5 × 10–41 | 39 (0 to 69) | 1.00 | <10–3 | AAA |
| SMAD7‡ | rs12953717 | 33 771 vs 32 364 (11‡) | 11 | 1.11 (1.07 to 1.15) | 1.4 × 10–7 | 46 (0 to 73) | 1.00 | 11 | 1.23 (1.16 to 1.29) | 6.0 × 10-13 | 36 (0 to 68) | 1.00 | 0.35 | ABA |
| SMAD7‡ | rs4464148 | 15 999 vs 15 216 (7||) | 7 | 1.14 (1.08 to 1.19) | 4.9 × 10–7 | 6 (0 to 72) | 1.00 | 7 | 1.30 (1.21 to 1.40) | 7.5 × 10-13 | 0 (0 to 71) | 1.00 | <10–3 | AAA |
| 8q24‡ | rs6983267 | 40 604 vs 42 672 (19) | 19 | 1.23 (1.18 to 1.27) | 7.4 × 10–30 | 38 (0 to 64) | 1.00 | 19 | 1.45 (1.39 to 1.51) | 6.9 × 10–68 | 0 (0 to 49) | 1.00 | <10–3 | ABA |
| 8q24¶¶ | rs10505477 | 18 580 vs 20 147(14) | 14 | 1.21 (1.15 to 1.28) | 1.0 × 10-13 | 25 (0 to 60) | 1.00 | 14 | 1.33 (1.26 to 1.41) | 6.2 × 10-23 | 44 (0 to 70) | 1.00 | <10–3 | ABA |
| 9p24¶¶ | rs719725 | 13 290 vs 14 774 (13) | 13 | 1.08 (1.00 to 1.16) | .04 | 0 (0 to 57) | 0.57 | 13 | 1.15 (1.07 to 1.24) | .0002 | 0 (0 to 57) | 0.96 | 0.93 | BAA |
| 19q13.1‡ | rs10411210 | 25 607 vs 26 477 (17) | 17 | 0.87 (0.81 to 0.93) | 7.8 × 10-5 | 55 (23 to 74) | 1.00 | 17 | 0.81 (0.70 to 0.93) | .003 | 0 (0 to 51) | 0.89 | 0.03 | ABA |
| 16q22.1‡ | rs9929218 | 26 191 vs 27 409 (18) | 18 | 0.93 (0.90 to 0.97) | .0001 | 13 (0 to 49) | 0.98 | 18 | 0.84 (0.78 to 0.90) | 1.9 × 10–7 | 0 (0 to 50) | 1.00 | 0.39 | AAA |
| 15q14¶¶ | rs4779584 | 13 656 vs 12 635 (9) | 9 | 1.13 (1.02 to 1.24) | .02 | 51 (0 to 77) | 0.99 | 9 | 1.38 (1.09 to 1.73) | .006 | 71 (47 to 85) | 1.00 | 0.83 | ABA |
| 1q41‡ | rs6691170 | 17 740 vs 19 776 (11) | 11 | 1.12 (1.07 to 1.17) | 2.9 × 10–7 | 19 (0 to 59) | 1.00 | 11 | 1.19 (1.12 to 1.27) | 1.2 × 10–7 | 0 (0 to 60) | 1.00 | <10–3 | AAA |
| 3q26.2‡ | rs10936599 | 17 802 vs 19 795 (11) | 11 | 0.90 (0.86 to 0.94) | 7.1 × 10–7 | 33 (0 to 67) | 1.00 | 11 | 0.85 (0.78 to 0.93) | .0004 | 0 (0 to 60) | 0.95 | 0.003 | ABA |
| 12q13.13¶¶ | rs11169552 | 17 148 vs 19 739 (11) | 11 | 0.92 (0.88 to 0.96) | .0003 | 0 (0 to 60) | 0.97 | 11 | 0.75 (0.66 to 0.86) | 1.2 × 10-5 | 53 (7 to 76) | 1.00 | 0.11 | AAA |
| 20q13.33‡ | rs4925386 | 17 847 vs 19 832 (11) | 11 | 0.91 (0.87 to 0.95) | 2.0 × 10-5 | 0 (0 to 60) | 0.99 | 11 | 0.80 (0.75 to 0.87) | 6.2 × 10-9 | 20 (0 to 59) | 1.00 | 0.02 | AAA |
| 14q22.2‡ | rs4444235 | 18 607 vs 19 576 (13) | 13 | 1.09 (1.04 to 1.14) | .0004 | 12 (0 to 52) | 0.95 | 13 | 1.18 (1.12 to 1.25) | 1.3 × 10–8 | 21 (0 to 59) | 1.00 | 0.14 | AAA |
| 20p12.3‡ | rs961253 | 18 118 vs 19 006 (13) | 13 | 1.13 (1.08 to 1.18) | 1.6 × 10–7 | 4 (0 to 58) | 1.00 | 13 | 1.22 (1.14 to 1.30) | 2.3 × 10–8 | 23 (0 to 60) | 1.00 | <10–3 | AAA |
| 8q23.3‡ | rs16892766 | 17 180 vs 17 840 (4)# | 4 | 1.27 (1.20 to 1.33) | 9.0 × 10-20 | 0 (0 to 85) | 1.00 | 4 | 1.38 (1.12 to 1.71) | .003 | 0 (0 to 85) | 0.90 | <10–3 | AAA |
| 10p14‡ | rs10795668 | 20 026 vs 20 682 (6)# | 6 | 0.89 (0.80 to 0.99) | .04 | 68 (23 to 86) | 0.57 | 6 | 0.77 (0.72 to 0.82) | 6.2 × 10-15 | 40 (0 to 76) | 1.00 | 0.90 | BCA |
| 11q23.1‡ | rs3802842 | 33 004 vs 31 654 (14) | 14 | 1.15 (1.12 to 1.19) | 1.2 × 10-17 | 33 (0 to 64) | 1.00 | 14 | 1.29 (1.23 to 1.36) | 1.310–20 | 33 (0 to 64) | 1.00 | <10–3 | ABA |
| Gene . | Variant . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | BFDP* . | Venice criteria grade† . | |||
| Common, low penetrance | ||||||||||||||
| SMAD7‡ | rs4939827 | 37 650 vs 36 154 (13§) | 13 | 0.89 (0.86 to 0.92) | 3.7 × 10-12 | 0 (0 to 57) | 1.00 | 13 | 0.75 (0.71 to 0.79) | 3.5 × 10–41 | 39 (0 to 69) | 1.00 | <10–3 | AAA |
| SMAD7‡ | rs12953717 | 33 771 vs 32 364 (11‡) | 11 | 1.11 (1.07 to 1.15) | 1.4 × 10–7 | 46 (0 to 73) | 1.00 | 11 | 1.23 (1.16 to 1.29) | 6.0 × 10-13 | 36 (0 to 68) | 1.00 | 0.35 | ABA |
| SMAD7‡ | rs4464148 | 15 999 vs 15 216 (7||) | 7 | 1.14 (1.08 to 1.19) | 4.9 × 10–7 | 6 (0 to 72) | 1.00 | 7 | 1.30 (1.21 to 1.40) | 7.5 × 10-13 | 0 (0 to 71) | 1.00 | <10–3 | AAA |
| 8q24‡ | rs6983267 | 40 604 vs 42 672 (19) | 19 | 1.23 (1.18 to 1.27) | 7.4 × 10–30 | 38 (0 to 64) | 1.00 | 19 | 1.45 (1.39 to 1.51) | 6.9 × 10–68 | 0 (0 to 49) | 1.00 | <10–3 | ABA |
| 8q24¶¶ | rs10505477 | 18 580 vs 20 147(14) | 14 | 1.21 (1.15 to 1.28) | 1.0 × 10-13 | 25 (0 to 60) | 1.00 | 14 | 1.33 (1.26 to 1.41) | 6.2 × 10-23 | 44 (0 to 70) | 1.00 | <10–3 | ABA |
| 9p24¶¶ | rs719725 | 13 290 vs 14 774 (13) | 13 | 1.08 (1.00 to 1.16) | .04 | 0 (0 to 57) | 0.57 | 13 | 1.15 (1.07 to 1.24) | .0002 | 0 (0 to 57) | 0.96 | 0.93 | BAA |
| 19q13.1‡ | rs10411210 | 25 607 vs 26 477 (17) | 17 | 0.87 (0.81 to 0.93) | 7.8 × 10-5 | 55 (23 to 74) | 1.00 | 17 | 0.81 (0.70 to 0.93) | .003 | 0 (0 to 51) | 0.89 | 0.03 | ABA |
| 16q22.1‡ | rs9929218 | 26 191 vs 27 409 (18) | 18 | 0.93 (0.90 to 0.97) | .0001 | 13 (0 to 49) | 0.98 | 18 | 0.84 (0.78 to 0.90) | 1.9 × 10–7 | 0 (0 to 50) | 1.00 | 0.39 | AAA |
| 15q14¶¶ | rs4779584 | 13 656 vs 12 635 (9) | 9 | 1.13 (1.02 to 1.24) | .02 | 51 (0 to 77) | 0.99 | 9 | 1.38 (1.09 to 1.73) | .006 | 71 (47 to 85) | 1.00 | 0.83 | ABA |
| 1q41‡ | rs6691170 | 17 740 vs 19 776 (11) | 11 | 1.12 (1.07 to 1.17) | 2.9 × 10–7 | 19 (0 to 59) | 1.00 | 11 | 1.19 (1.12 to 1.27) | 1.2 × 10–7 | 0 (0 to 60) | 1.00 | <10–3 | AAA |
| 3q26.2‡ | rs10936599 | 17 802 vs 19 795 (11) | 11 | 0.90 (0.86 to 0.94) | 7.1 × 10–7 | 33 (0 to 67) | 1.00 | 11 | 0.85 (0.78 to 0.93) | .0004 | 0 (0 to 60) | 0.95 | 0.003 | ABA |
| 12q13.13¶¶ | rs11169552 | 17 148 vs 19 739 (11) | 11 | 0.92 (0.88 to 0.96) | .0003 | 0 (0 to 60) | 0.97 | 11 | 0.75 (0.66 to 0.86) | 1.2 × 10-5 | 53 (7 to 76) | 1.00 | 0.11 | AAA |
| 20q13.33‡ | rs4925386 | 17 847 vs 19 832 (11) | 11 | 0.91 (0.87 to 0.95) | 2.0 × 10-5 | 0 (0 to 60) | 0.99 | 11 | 0.80 (0.75 to 0.87) | 6.2 × 10-9 | 20 (0 to 59) | 1.00 | 0.02 | AAA |
| 14q22.2‡ | rs4444235 | 18 607 vs 19 576 (13) | 13 | 1.09 (1.04 to 1.14) | .0004 | 12 (0 to 52) | 0.95 | 13 | 1.18 (1.12 to 1.25) | 1.3 × 10–8 | 21 (0 to 59) | 1.00 | 0.14 | AAA |
| 20p12.3‡ | rs961253 | 18 118 vs 19 006 (13) | 13 | 1.13 (1.08 to 1.18) | 1.6 × 10–7 | 4 (0 to 58) | 1.00 | 13 | 1.22 (1.14 to 1.30) | 2.3 × 10–8 | 23 (0 to 60) | 1.00 | <10–3 | AAA |
| 8q23.3‡ | rs16892766 | 17 180 vs 17 840 (4)# | 4 | 1.27 (1.20 to 1.33) | 9.0 × 10-20 | 0 (0 to 85) | 1.00 | 4 | 1.38 (1.12 to 1.71) | .003 | 0 (0 to 85) | 0.90 | <10–3 | AAA |
| 10p14‡ | rs10795668 | 20 026 vs 20 682 (6)# | 6 | 0.89 (0.80 to 0.99) | .04 | 68 (23 to 86) | 0.57 | 6 | 0.77 (0.72 to 0.82) | 6.2 × 10-15 | 40 (0 to 76) | 1.00 | 0.90 | BCA |
| 11q23.1‡ | rs3802842 | 33 004 vs 31 654 (14) | 14 | 1.15 (1.12 to 1.19) | 1.2 × 10-17 | 33 (0 to 64) | 1.00 | 14 | 1.29 (1.23 to 1.36) | 1.310–20 | 33 (0 to 64) | 1.00 | <10–3 | ABA |
|| Includes unpublished data from Scottish GWAS.
* Bayesian False Discovery Probability (BFDP) value for the heterozygous additive model (var/wt vs wt/wt) at prior probability of 0.05. BFDP level of noteworthiness 0.2. For more information, please see Supplementary Table 6.
† Venice criteria grade for the heterozygous additive model (var/wt vs wt/wt). For the third criterion (protection from bias), all meta-analyses of variants identified by GWAS were scored with A. Reporting was generally more transparent (at least for the discovery datasets, but somewhat variable for the replication datasets). All of the studies included the discovery data with the replication data in estimating magnitude of effect and arguably this may bias the magnitude of the association. However, it is unlikely to affect direction of association. Therefore, we rated the third category for GWASs as “A.”
# Tomlinson 2008 was based on 10 samples.
§ Includes unpublished data from Canadian GWAS.
‡ The associations considered to be “positives”.
¶¶ The associations considered to be “less-credible positives”.
Summary crude odds ratios (ORs) and 95% confidence intervals (95% CIs) for two additive models for variants that were identified from genome-wide association studies
| Gene . | Variant . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | BFDP* . | Venice criteria grade† . | |||
| Common, low penetrance | ||||||||||||||
| SMAD7‡ | rs4939827 | 37 650 vs 36 154 (13§) | 13 | 0.89 (0.86 to 0.92) | 3.7 × 10-12 | 0 (0 to 57) | 1.00 | 13 | 0.75 (0.71 to 0.79) | 3.5 × 10–41 | 39 (0 to 69) | 1.00 | <10–3 | AAA |
| SMAD7‡ | rs12953717 | 33 771 vs 32 364 (11‡) | 11 | 1.11 (1.07 to 1.15) | 1.4 × 10–7 | 46 (0 to 73) | 1.00 | 11 | 1.23 (1.16 to 1.29) | 6.0 × 10-13 | 36 (0 to 68) | 1.00 | 0.35 | ABA |
| SMAD7‡ | rs4464148 | 15 999 vs 15 216 (7||) | 7 | 1.14 (1.08 to 1.19) | 4.9 × 10–7 | 6 (0 to 72) | 1.00 | 7 | 1.30 (1.21 to 1.40) | 7.5 × 10-13 | 0 (0 to 71) | 1.00 | <10–3 | AAA |
| 8q24‡ | rs6983267 | 40 604 vs 42 672 (19) | 19 | 1.23 (1.18 to 1.27) | 7.4 × 10–30 | 38 (0 to 64) | 1.00 | 19 | 1.45 (1.39 to 1.51) | 6.9 × 10–68 | 0 (0 to 49) | 1.00 | <10–3 | ABA |
| 8q24¶¶ | rs10505477 | 18 580 vs 20 147(14) | 14 | 1.21 (1.15 to 1.28) | 1.0 × 10-13 | 25 (0 to 60) | 1.00 | 14 | 1.33 (1.26 to 1.41) | 6.2 × 10-23 | 44 (0 to 70) | 1.00 | <10–3 | ABA |
| 9p24¶¶ | rs719725 | 13 290 vs 14 774 (13) | 13 | 1.08 (1.00 to 1.16) | .04 | 0 (0 to 57) | 0.57 | 13 | 1.15 (1.07 to 1.24) | .0002 | 0 (0 to 57) | 0.96 | 0.93 | BAA |
| 19q13.1‡ | rs10411210 | 25 607 vs 26 477 (17) | 17 | 0.87 (0.81 to 0.93) | 7.8 × 10-5 | 55 (23 to 74) | 1.00 | 17 | 0.81 (0.70 to 0.93) | .003 | 0 (0 to 51) | 0.89 | 0.03 | ABA |
| 16q22.1‡ | rs9929218 | 26 191 vs 27 409 (18) | 18 | 0.93 (0.90 to 0.97) | .0001 | 13 (0 to 49) | 0.98 | 18 | 0.84 (0.78 to 0.90) | 1.9 × 10–7 | 0 (0 to 50) | 1.00 | 0.39 | AAA |
| 15q14¶¶ | rs4779584 | 13 656 vs 12 635 (9) | 9 | 1.13 (1.02 to 1.24) | .02 | 51 (0 to 77) | 0.99 | 9 | 1.38 (1.09 to 1.73) | .006 | 71 (47 to 85) | 1.00 | 0.83 | ABA |
| 1q41‡ | rs6691170 | 17 740 vs 19 776 (11) | 11 | 1.12 (1.07 to 1.17) | 2.9 × 10–7 | 19 (0 to 59) | 1.00 | 11 | 1.19 (1.12 to 1.27) | 1.2 × 10–7 | 0 (0 to 60) | 1.00 | <10–3 | AAA |
| 3q26.2‡ | rs10936599 | 17 802 vs 19 795 (11) | 11 | 0.90 (0.86 to 0.94) | 7.1 × 10–7 | 33 (0 to 67) | 1.00 | 11 | 0.85 (0.78 to 0.93) | .0004 | 0 (0 to 60) | 0.95 | 0.003 | ABA |
| 12q13.13¶¶ | rs11169552 | 17 148 vs 19 739 (11) | 11 | 0.92 (0.88 to 0.96) | .0003 | 0 (0 to 60) | 0.97 | 11 | 0.75 (0.66 to 0.86) | 1.2 × 10-5 | 53 (7 to 76) | 1.00 | 0.11 | AAA |
| 20q13.33‡ | rs4925386 | 17 847 vs 19 832 (11) | 11 | 0.91 (0.87 to 0.95) | 2.0 × 10-5 | 0 (0 to 60) | 0.99 | 11 | 0.80 (0.75 to 0.87) | 6.2 × 10-9 | 20 (0 to 59) | 1.00 | 0.02 | AAA |
| 14q22.2‡ | rs4444235 | 18 607 vs 19 576 (13) | 13 | 1.09 (1.04 to 1.14) | .0004 | 12 (0 to 52) | 0.95 | 13 | 1.18 (1.12 to 1.25) | 1.3 × 10–8 | 21 (0 to 59) | 1.00 | 0.14 | AAA |
| 20p12.3‡ | rs961253 | 18 118 vs 19 006 (13) | 13 | 1.13 (1.08 to 1.18) | 1.6 × 10–7 | 4 (0 to 58) | 1.00 | 13 | 1.22 (1.14 to 1.30) | 2.3 × 10–8 | 23 (0 to 60) | 1.00 | <10–3 | AAA |
| 8q23.3‡ | rs16892766 | 17 180 vs 17 840 (4)# | 4 | 1.27 (1.20 to 1.33) | 9.0 × 10-20 | 0 (0 to 85) | 1.00 | 4 | 1.38 (1.12 to 1.71) | .003 | 0 (0 to 85) | 0.90 | <10–3 | AAA |
| 10p14‡ | rs10795668 | 20 026 vs 20 682 (6)# | 6 | 0.89 (0.80 to 0.99) | .04 | 68 (23 to 86) | 0.57 | 6 | 0.77 (0.72 to 0.82) | 6.2 × 10-15 | 40 (0 to 76) | 1.00 | 0.90 | BCA |
| 11q23.1‡ | rs3802842 | 33 004 vs 31 654 (14) | 14 | 1.15 (1.12 to 1.19) | 1.2 × 10-17 | 33 (0 to 64) | 1.00 | 14 | 1.29 (1.23 to 1.36) | 1.310–20 | 33 (0 to 64) | 1.00 | <10–3 | ABA |
| Gene . | Variant . | Case vs control subjects (no. of samples) . | Additive Model: var/wt vs wt/wt . | Additive Model: var/var vs wt/wt . | Credibility . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | N . | OR (95% CI) . | P . | I2 (95% CI) . | Power . | BFDP* . | Venice criteria grade† . | |||
| Common, low penetrance | ||||||||||||||
| SMAD7‡ | rs4939827 | 37 650 vs 36 154 (13§) | 13 | 0.89 (0.86 to 0.92) | 3.7 × 10-12 | 0 (0 to 57) | 1.00 | 13 | 0.75 (0.71 to 0.79) | 3.5 × 10–41 | 39 (0 to 69) | 1.00 | <10–3 | AAA |
| SMAD7‡ | rs12953717 | 33 771 vs 32 364 (11‡) | 11 | 1.11 (1.07 to 1.15) | 1.4 × 10–7 | 46 (0 to 73) | 1.00 | 11 | 1.23 (1.16 to 1.29) | 6.0 × 10-13 | 36 (0 to 68) | 1.00 | 0.35 | ABA |
| SMAD7‡ | rs4464148 | 15 999 vs 15 216 (7||) | 7 | 1.14 (1.08 to 1.19) | 4.9 × 10–7 | 6 (0 to 72) | 1.00 | 7 | 1.30 (1.21 to 1.40) | 7.5 × 10-13 | 0 (0 to 71) | 1.00 | <10–3 | AAA |
| 8q24‡ | rs6983267 | 40 604 vs 42 672 (19) | 19 | 1.23 (1.18 to 1.27) | 7.4 × 10–30 | 38 (0 to 64) | 1.00 | 19 | 1.45 (1.39 to 1.51) | 6.9 × 10–68 | 0 (0 to 49) | 1.00 | <10–3 | ABA |
| 8q24¶¶ | rs10505477 | 18 580 vs 20 147(14) | 14 | 1.21 (1.15 to 1.28) | 1.0 × 10-13 | 25 (0 to 60) | 1.00 | 14 | 1.33 (1.26 to 1.41) | 6.2 × 10-23 | 44 (0 to 70) | 1.00 | <10–3 | ABA |
| 9p24¶¶ | rs719725 | 13 290 vs 14 774 (13) | 13 | 1.08 (1.00 to 1.16) | .04 | 0 (0 to 57) | 0.57 | 13 | 1.15 (1.07 to 1.24) | .0002 | 0 (0 to 57) | 0.96 | 0.93 | BAA |
| 19q13.1‡ | rs10411210 | 25 607 vs 26 477 (17) | 17 | 0.87 (0.81 to 0.93) | 7.8 × 10-5 | 55 (23 to 74) | 1.00 | 17 | 0.81 (0.70 to 0.93) | .003 | 0 (0 to 51) | 0.89 | 0.03 | ABA |
| 16q22.1‡ | rs9929218 | 26 191 vs 27 409 (18) | 18 | 0.93 (0.90 to 0.97) | .0001 | 13 (0 to 49) | 0.98 | 18 | 0.84 (0.78 to 0.90) | 1.9 × 10–7 | 0 (0 to 50) | 1.00 | 0.39 | AAA |
| 15q14¶¶ | rs4779584 | 13 656 vs 12 635 (9) | 9 | 1.13 (1.02 to 1.24) | .02 | 51 (0 to 77) | 0.99 | 9 | 1.38 (1.09 to 1.73) | .006 | 71 (47 to 85) | 1.00 | 0.83 | ABA |
| 1q41‡ | rs6691170 | 17 740 vs 19 776 (11) | 11 | 1.12 (1.07 to 1.17) | 2.9 × 10–7 | 19 (0 to 59) | 1.00 | 11 | 1.19 (1.12 to 1.27) | 1.2 × 10–7 | 0 (0 to 60) | 1.00 | <10–3 | AAA |
| 3q26.2‡ | rs10936599 | 17 802 vs 19 795 (11) | 11 | 0.90 (0.86 to 0.94) | 7.1 × 10–7 | 33 (0 to 67) | 1.00 | 11 | 0.85 (0.78 to 0.93) | .0004 | 0 (0 to 60) | 0.95 | 0.003 | ABA |
| 12q13.13¶¶ | rs11169552 | 17 148 vs 19 739 (11) | 11 | 0.92 (0.88 to 0.96) | .0003 | 0 (0 to 60) | 0.97 | 11 | 0.75 (0.66 to 0.86) | 1.2 × 10-5 | 53 (7 to 76) | 1.00 | 0.11 | AAA |
| 20q13.33‡ | rs4925386 | 17 847 vs 19 832 (11) | 11 | 0.91 (0.87 to 0.95) | 2.0 × 10-5 | 0 (0 to 60) | 0.99 | 11 | 0.80 (0.75 to 0.87) | 6.2 × 10-9 | 20 (0 to 59) | 1.00 | 0.02 | AAA |
| 14q22.2‡ | rs4444235 | 18 607 vs 19 576 (13) | 13 | 1.09 (1.04 to 1.14) | .0004 | 12 (0 to 52) | 0.95 | 13 | 1.18 (1.12 to 1.25) | 1.3 × 10–8 | 21 (0 to 59) | 1.00 | 0.14 | AAA |
| 20p12.3‡ | rs961253 | 18 118 vs 19 006 (13) | 13 | 1.13 (1.08 to 1.18) | 1.6 × 10–7 | 4 (0 to 58) | 1.00 | 13 | 1.22 (1.14 to 1.30) | 2.3 × 10–8 | 23 (0 to 60) | 1.00 | <10–3 | AAA |
| 8q23.3‡ | rs16892766 | 17 180 vs 17 840 (4)# | 4 | 1.27 (1.20 to 1.33) | 9.0 × 10-20 | 0 (0 to 85) | 1.00 | 4 | 1.38 (1.12 to 1.71) | .003 | 0 (0 to 85) | 0.90 | <10–3 | AAA |
| 10p14‡ | rs10795668 | 20 026 vs 20 682 (6)# | 6 | 0.89 (0.80 to 0.99) | .04 | 68 (23 to 86) | 0.57 | 6 | 0.77 (0.72 to 0.82) | 6.2 × 10-15 | 40 (0 to 76) | 1.00 | 0.90 | BCA |
| 11q23.1‡ | rs3802842 | 33 004 vs 31 654 (14) | 14 | 1.15 (1.12 to 1.19) | 1.2 × 10-17 | 33 (0 to 64) | 1.00 | 14 | 1.29 (1.23 to 1.36) | 1.310–20 | 33 (0 to 64) | 1.00 | <10–3 | ABA |
|| Includes unpublished data from Scottish GWAS.
* Bayesian False Discovery Probability (BFDP) value for the heterozygous additive model (var/wt vs wt/wt) at prior probability of 0.05. BFDP level of noteworthiness 0.2. For more information, please see Supplementary Table 6.
† Venice criteria grade for the heterozygous additive model (var/wt vs wt/wt). For the third criterion (protection from bias), all meta-analyses of variants identified by GWAS were scored with A. Reporting was generally more transparent (at least for the discovery datasets, but somewhat variable for the replication datasets). All of the studies included the discovery data with the replication data in estimating magnitude of effect and arguably this may bias the magnitude of the association. However, it is unlikely to affect direction of association. Therefore, we rated the third category for GWASs as “A.”
# Tomlinson 2008 was based on 10 samples.
§ Includes unpublished data from Canadian GWAS.
‡ The associations considered to be “positives”.
¶¶ The associations considered to be “less-credible positives”.
To assess the credibility of genetic associations, we considered the Venice criteria (11,12) and the BFDP (26) (Supplementary Table 6, available online). Applying these filters indicate that associations with 16 variants (17% of the meta-analyzed SNPs) tagging 13 loci (SMAD7, MUTYH, MTHFR, and the 8q24, 8q23.3, 11q23.1, 14q22.2, 1q41, 20p12.3, 20q13.33, 3q26.2, 16q22.1, and 19q13.1) represent the most credible findings and these will be referred as “positive” SNP associations (Tables 2 and 3; Supplementary Tables 2 and 3 and Supplementary Figures 1–64, available on the CRCgene website). These findings are based on accrued data on 16 000–40 604 case patients and on 15 216–42 672 control subjects, with a median of 1616 case patients per study. All variants that were included in the “positive” SNP associations reached genome-wide statistical significance (P ≤ 1.7 × 10–7) in at least one meta-analysis model, apart from the ones in MUTYH and in 19q13.1.
We identified “less-credible positive” associations (higher heterogeneity, lower statistical power, BFDP > 0.2) with 23 variants (25% of the meta-analyzed SNPs) of 22 loci (Tables 2 and 3; Supplementary Tables 2 and 3 and Supplementary Figures 65–146, available on the CRCgene website. These findings were based on accrued data on 857–26 370 case patients and on 932–26 662 control subjects, with a median of at least 600 case patients per study. Variants in the 10p14 and 15q14 loci reached genome-wide statistical significance (P ≤ 1.7 × 10–7) in at least one meta-analysis model.
For those SNPs that were identified as “positives” or “less-credible positives” after applying the BFDP and Venice criteria, we applied the model-free meta-analysis approach as described by Thompson et al. (32) (Supplementary Table 7, available online), which gives an estimate, λ, of the underlying genetic model. Funnel plots were produced for all the positive and less-credible associations. There was no evidence of small-study effects, apart from the associations with rs1799750 (MMP1), rs34743033 (TS), GSTM1 deletion, rs36053993 (MUTYH), rs10411210 (19q13.1), rs10936599 (3q26.2). However, only the GSTM1 deletion and rs10411210 (19q13.1) and rs10936599 (3q26.2) tests were based on more than 10 studies and satisfied the conditions of no heterogeneity. Therefore, the results of the other SNPs, for which there was some evidence of a small-study effect, should be interpreted with caution. The remaining 53 (58%) meta-analyzed SNPs of 37 loci are designated as “negatives,” based on accrued data from 575 to 17 178 case patients and 836 to 24 792 control subjects, with an average of at least 600 case patients per study.
Discussion
This is the first systematic, comprehensive field synopsis of genetic association studies on colorectal cancer to our knowledge. It differs from the recently published database of cancer genetic association (33) in that it has collated and extracted data from more than 600 publications on more than 400 polymorphisms in 110 different genes and presents the results of more than 90 new meta-analyses, rather than acting as a portal to already published meta-analyses and GWASs. Furthermore, we have classified the results of our analysis as “positive associations,” “less-credible positive associations,” and “negative associations” within a defined statistical and causal inference framework, including the Venice criteria (11,12) the BDFP, and the model-free meta-analysis approach.
Two SNPs at the 8q24 locus (rs6983267 and rs10505477) identified in the GWASs had a large volume of evidence and showed positive associations irrespective of the genetic model considered, with moderate heterogeneity, and the model-free approach suggested an underlying additive model (Table 3, Supplementary Tables 3 and 7). The SNP rs6983267 may be a somatic target in colorectal cancer (34) and may be associated with enhanced responsiveness to Wnt signaling (35). Further, rs6983267 has also been found to be associated with other types of cancer (36,37), including prostate cancer (38–40). Interaction with the MYC proto-oncogene has been controversial (41–45), but recently in functional studies in cell lines, interaction between enhancer elements in the 8q24 locus and the MYC promoter, via transcription factor Tcf-4 binding and allele-specific regulation of MYC expression, has been demonstrated (46).
One variant close to SMAD7 (rs4939827), which was inversely associated with colorectal cancer, and two variants (rs12953717 and rs4464148), which were associated with increased risk for the disease, were again identified through GWASs with large numbers of participants. The associations were apparent irrespective of the genetic model, with moderate heterogeneity, and the model-free approach suggested an underlying additive model (Table 3, Supplementary Tables 3 and 7). The SMAD7 protein is an inhibitor for the TGFβ signaling pathway (47). The otherwise-tentative associations with the TGFβ1 rs1800469 (C5097) and TGFβR1 rs11466445 (*6A) variants may be consistent with this finding.
Dysfunction of base-excision repair, the major pathway for repairing oxidative damage, has been implicated in the development of multiple colorectal adenomas and colorectal cancer (MUTYH-associated polyposis [MAP] syndrome). Two variants of MUTYH were associated with increased risk of colorectal cancer, with little heterogeneity apparent (Table 2, Supplementary Table 2), providing further evidence that the single variants can act as lower-penetrance risk alleles of smaller effect, validating the recent results for Y179C (rs34612342) (48) and also highlighting the possible role of G396D (rs36053993). The BFDP was substantially less for rs36053993 than for rs34612342, possibly because of the very low MAF of the rs34612342 variant, and therefore, rs36053993 was classified as having a positive association, whereas rs34612342 was classified as having a less-credible positive association with colorectal cancer (Supplementary Table 6). Although there was formally statistically significant evidence of small-study effects for rs36053993 (Supplementary Figures 189–190, available on the CRCgene website), the magnitude of this effect was small and has to be interpreted in the context of multiple tests for small-study effects. The model-free approach suggested a recessive model for rs36053993 and an additive (but with wide CI) for rs34612342 (Supplementary Table 7).
An inverse association with homozygosity for the MTHFR rs1801133 (commonly reported as C677T) variant was observed in 52 studies comparing more than 27 000 case patients and almost 40 000 control subjects, with moderate heterogeneity and an underlying recessive model as suggested by the model-free approach (Table 2, Supplementary Tables 2 and 7). A similar association was observed in a smaller body of evidence with the rs1801131 (commonly known as A1298C) variant, with little heterogeneity between 34 studies (Table 2, Supplementary Table 4). In most populations, strong LD between the variants has been observed (49,50). These findings extend and concur with previous systematic reviews and meta-analyses (51–56). All of the studies considered these MTHFR variants as candidates for association with colorectal cancer on the basis of knowledge about their effects on enzyme function and associations with blood levels of folate and related metabolites. However, the evidence is further reinforced by GWASs of plasma homocysteine confirming some of the earlier work on the effects of the MTHFR C677T variant (57–60).
The other variants with which associations were observed were all based on accumulated data on a total of at least 17 000 case patients and 18 000 control subjects, with a median of 1506 case patients per study. Positive associations with colorectal cancer were identified with variants at the 8q23.3 (rs16892766), 11q23.1 (rs3802842), 14q22.2 (rs4444235), 1q41 (rs6691170), and 20p12.3 (rs961253) loci, and inverse associations were identified with variants at the 20q13.33 (rs4925386), 3q26.2 (rs10936599), 16q22.1 (rs9929218), and 19q13.1 (rs10411210) loci. Although there was formally statistically significant evidence of small-study effects for 3q26.2 (rs10936599) (Supplementary Figures 209–210, available on the CRCgene website) and 19q13.1 (rs10411210; Supplementary Figures 203–204), the magnitude of these effects was not large, and the findings of GWASs are less prone to the problem of selective reporting than studies that target only a few risk factors at a time with only selected findings being published (61). In a study of somatic genetic changes, no allelic imbalance targeting 11q23.1 (rs3802842), 14q22.2 (rs4444235), 16q22.1 (rs9929218), 19q13.1 (rs10411210), or 20p12.3 (rs961253) was observed (Table 3, Supplementary Table 3) (62).
The less-credible positive associations with 23 variants of 22 genes involved the following pathways—adhesion (CDH1, MMP1, and MMP3); alcohol metabolism (ALDH2); base-excision repair (XRCC1 and MUTYH); inflammation and immune response (IL6, two variants of NOD2); inhibition of cell growth (CCND1, TGFB1, and TGFBR1); one-carbon metabolism (TS); substrate metabolism (CYP1A1, GSTM1, GSTT1, and NAT1); vitamin D metabolism (VDR)—common low-penetrance variants at 9p24 (rs719725), 10p14 (rs10795668), 15q14 (rs4779584), and 12q13.13 (rs11169552); and the common rs459552 (D1822V) variant of APC, for which rare variants confer a high risk of colorectal cancer. Some of these have been considered in previous meta-analyses.
The association with the CDH1 rs16260 (C160A) variant remained apparent when the analysis was restricted to populations of European origin (Supplementary Tables 4 and 5). No association was apparent in an earlier meta-analysis by Wang et al. (63), but this meta-analysis included less than one-tenth of the number of case and control subjects included in the present study. Previous meta-analyses have found positive associations with the MMP1 rs1799750 (G-1607GG) but not the MMP3 rs3025058 (AAAAA 1612 AAAAAA) variant (64,65). We found evidence of small-study effects for MMP1 rs1799750 (Supplementary Figures 149–150, available on the CRCgene website.
With regard to ALDH2, our findings are similar to those of Wang et al. (66). All of these studies were carried out in Asian populations, in which the rs671 (Glu487Lys) variant is more common than in the other populations (MAF is 0 in white, 0.16 in Chinese, and 0.23 in Japanese populations). The association between the ALDH2 variant and colorectal adenomas has been investigated, but it has been interpreted as null (67,68).
Inverse associations were observed with the XRCC1 rs25487 (Arg399Gln) variant in the additive and recessive models with heterogeneity, which persisted when the analysis was restricted to populations of European origin (Table 2, Supplementary Tables 2, 4, and 5). An earlier meta-analysis with about half the number of participants also detected an inverse association for the recessive model (69), but another meta-analysis that included a smaller number of participants (but more studies) was interpreted as null (70). It has been suggested that the association between this XRCC1 variant and colorectal adenomas is stronger than that apparent with colorectal cancer (71), but we note that the volume of evidence relating to adenomas is substantially smaller. Three variants of NOD1 (rs5743293, rs2066845, and rs2066844) are associated with increased risk when a dominant model is assumed (Supplementary Table 2); this finding is consistent with that of Tian et al. (72).
Assuming a dominant model, variants at three gene loci that are involved in the inhibition of cell growth are associated with an increased risk of colorectal cancer. First, the analysis of the CCND1 rs17852153 (G870A) variant includes approximately five times as many participants as the earlier study of Tan et al. (73) (Supplementary Table 4). In the study of Tan et al., there was a concern about small-study effects, but we found no evidence of this problem (Supplementary Figures 165–166). Two studies of colorectal adenomas give inconsistent results (74,75). Second, the TGFB1 rs1800469 (C5097) variant has been investigated solely in Asian populations (Supplementary Table 2) (76). Third, previous meta-analyses have drawn different conclusions about the association with the TGFBR1 rs11466445 (*6A) variant (77–79). The magnitude of association in the present meta-analysis is similar to that determined in previous reports (78,79), but the inclusion of data on an additional 2000 participants from two studies has increased statistical power but not heterogeneity (Supplementary Table 2).
Our finding of an inverse association with the TS rs34743033 (TSER) variant assuming either an additive or dominant model remained apparent when the analysis was restricted to populations of European origin (Table 2, Supplementary Tables 2, 4, and 5), although there was evidence of small-study effects for the comparison between homozygotes under the additive model. Two studies of colorectal adenomas do not suggest an overall association with this variant (80,81), and one study did not detect an association with adenoma recurrence (82).
There has been a longstanding interest in the possible effects of genetic variants influencing the metabolism of diverse substrates, including potential carcinogens, on cancer risk. Cytochrome P-450 CYP1A1 is involved in the metabolism of polycyclic aromatic hydrocarbons and estrogens, and perhaps cruciferous vegetables and heterocyclic amines (83). Further, CYP1A1 mRNA has been detected in the large bowel (84,85), and human colon cell lines also express CYP1A1 at the protein and mRNA levels (86). These observations stimulated many candidate gene studies of the association between colorectal cancer and CYP1A1 variants. The positive association with presence of the rs1048943 (A2454G, Ile462Val) variant apparent in the present analysis is in line with the results of a smaller recent meta-analysis (87), whereas an earlier meta-analysis (88) did not detect this association (Table 2, Supplementary Table 2). No association was apparent with the rs4646903 variant, but there was low statistical power to detect this association (Table 2, Supplementary Table 2).
Another intensively investigated gene polymorphism that is thought to affect the metabolism of multiple substrates is the GSTM1 null variant, with which we observed a weak positive association in data from more than 18 000 case patients and more than 26 000 control subjects from 43 studies (Supplementary Table 2), with some evidence of small-study effects. The weak positive association was also apparent when analysis was restricted to 25 studies in populations of European origin (Supplementary Table 5). There have been several previous meta-analyses; the earliest of these studies indicated no overall association (88–90), whereas the more recent studies showed a positive association (91–93). No clear association between GSTM1 and adenomas has been observed (94). In data from more than 13 000 case patients and 20 000 control subjects from 35 studies, there was also a positive association with the GSTT1 null deletion, in line with previous meta-analyses (Supplementary Table 2) (88,92,95,96). The positive association remained apparent when analysis was restricted to approximately half the number of participants from 20 studies in populations of European origin (Supplementary Table 5). Studies of GSTT1 and adenomas have not detected a clear association (94).
In data from nearly 5000 case patients and more than 6000 control subjects from 15 studies, there was a positive association with heterogeneity for NAT1 alleles classified as being associated with rapid acetylation (Table 2, Supplementary Table 2). No association was apparent with homozygosity for alleles associated with rapid acetylation or assuming other models of inheritance. Furthermore, the association was not apparent when analysis was restricted to populations of European origin (Supplementary Tables 4 and 5). A previous meta-analysis found no association with carriage of the NAT1*10 variant in less than 1000 participants from three studies (88).
There was an inverse association between colorectal cancer and the VDR rs1544410 (Bsml 6089GA) variant, but with considerable heterogeneity between studies (Table 2, Supplementary Table 2). This heterogeneity was reduced when analysis was limited to populations of European origin (Supplementary Tables 4 and 5). No association between this variant and colorectal adenomas (97–99) or recurrence (100,101) has been observed.
On the basis of GWASs, a positive association with the rs719725 variant at 9p24 was observed with moderate heterogeneity in data from more than 11 000 case patients and 13 000 control subjects from 13 studies. This finding is consistent with the recent meta-analysis of Kocarnick et al. (102).
Assuming a recessive mode of inheritance, an inverse association was found with the rs10795668 at 10p14, based on data from more than 20 000 case patients and more than 20 000 control subjects from six studies (Table 3). The magnitude of association was attenuated when the analysis was restricted to populations of European origin (Supplementary Table 4). In a study of somatic genetic changes, no allelic imbalance targeting this SNP was observed (62).
We considered the associations with one variant at 15q14 (rs4779584) and one at 12q13.13 (rs11169552) to lie in the less-credible category of associations because of heterogeneity between studies. Based on data from more than 13 500 case patients and 12 500 control subjects from nine studies, there was a positive association with the rs4779584 variant (Table 3, Supplementary Table 3), and based on data from nearly 18 000 case patients and nearly 20 000 control subjects from 11 studies, an inverse association with the rs11169552 variant (Table 3, Supplementary Table 3). In a study of somatic genetic changes, no allelic imbalance targeting the former variant was observed (62).
On the basis of studies carried out solely in populations of European origin, homozygosity for the common rs459552 (D1822V) variant of APC, rare truncating germline variants of which cause familial adenomatous polyposis (FAP), was inversely associated with colorectal cancer (Supplementary Table 4). No marginal association between this variant and colorectal adenomas (103,104) or advanced adenomas (ie, ≥1cm or containing villous elements or high-grade dysplasia) of the distal colon (105) has been observed.
For variants for which we did not find associations in our meta-analyses, the accumulated number of case patients ranged between nearly 600 and slightly more than 15 000, with 20 variants in which the accumulated number of case patients exceeded 5000 (Table 2, Supplementary Table 2).
The lack of association with the PPARγ rs1801282 variant does not support an earlier meta-analysis (106), whereas the lack of association with the PPARγ rs3856606 variant is consistent with the results of Lu et al. (107). Earlier work on NAT2 suggested a positive association with the rapid acetylator phenotype, but no association with acetylator status could be inferred from genotype (51,88,108,109). No overall association was apparent in an earlier meta-analysis with homozygosity for the MTR rs1805087 (A2576G) variant, based on nine studies (110). The current analysis approximately doubles the data on colorectal cancer and the GSTP1 rs1695 (Ile105Val) variant. As in the meta-analysis of Economoupoulos and Sergentanis (92), it does not confirm an earlier analysis that found an inverse association when a recessive model was assumed (111). Studies of colorectal adenomas do not suggest any association with this GSTP1 variant, although subgroup effects have been reported (112–115).
The results presented here are in accordance with previous meta-analyses that did not show any association with the IGF1 CA-repeat polymorphism (116,117), although one of these showed an association with a higher serum concentration of IGF1 (117). Our finding of no association with the IGFBP3 rs2854744 (A202C) variant is consistent with that reported by Chen (117).
There have been numerous studies assessing the possible relationship between common variants of the TP53 tumor suppressor gene and several cancer types. The relationship between colorectal cancer and the rs1042522 (codon 72) variant has been investigated in previous meta-analyses, none of which suggest an association across multiple small studies, although marked heterogeneity was observed (118–121). Of note, one of these highlights the differences in results according to genotyping method and source of tissue for genotyping case patients (118). With regard to the APC rs1801166 (E1317Q) variant, one previous analysis of colorectal cancer was interpreted as indicating no association (122).
The current analysis examines more than double the amount of genetic association data for the XRCC1 variant (Arg194Trp) and is consistent with previous meta-analyses indicating no association (69,70).
Because of the inverse association between aspirin and colorectal neoplasia, many investigations have been carried out on genetic variants that might influence cyclooxygenase (COX) metabolism. We did not find any association with PTGS2/COX2 variants: neither with the rs20417 (G765C) variant (for which there is most evidence), in accordance with Cao (123), nor with the other variants. Individually, adenoma studies did not suggest associations for newly detected adenomas with the rs20417 (124–127), the rs5275 (124,125), the rs5277 (124,125), or the rs689466 variants (125,126). With regard to increased risk of adenoma recurrence, individual studies found no associations with the rs20417 (128,129) or the rs5275 variants (129), but one study reported an increased risk associated with homozygosity for the uncommon variant of rs5277 (129).
The lack of association with the NQ01 rs1800566 variant was also found in an earlier meta-analysis based on about one-third the number of subjects (130). With regard to the other variants for which we conducted meta-analyses, our finding of no association is consistent with earlier meta-analyses for MMP9 (64), VEGF rs2010963 (G634C) (131), XRCC1 rs25489 (Arg280His) (70), XRCC3 rs861359 (Thr241Met) (69), TNFα rs1800629 (308G>A) (132), and CYP2E1 rs3813867 (Pstl/Rsal, 1293G>C) (133).
We were not able to identify previous meta-analyses of ADH1B rs1229984, ADH1C rs698, VEGF rs3025039, OGG1 rs1052133, IL8 rs4073, IL10 rs1800896, APOE rs429358, TS rs34489327, CYP1A1 rs4646903, CYP1A2 rs762551, CYP2E1 rs2031920, GSTP1 rs1138272, VDR rs731236 or the MLH1 rs1799977, and rs1800734 variants.
The substantial majority of studies are from Western Europe, North America, and parts of Asia (more than one study from China, Japan, Korea, Singapore, and Taiwan). Investigation of more diverse populations is important, as it will enable variants to be considered in populations with more diverse patterns of LD and gene–environment interaction.
The number of common, low-penetrance variants that appear to be associated with colorectal cancer is very much less than anticipated, therefore decreasing the feasibility of combining variants as a profile in a prediction tool for stratifying screening modalities or primary prevention approaches (134). In addition, the variants so far identified account for only a small proportion of the familial risk (6.1% for “positive” and 14.1% when adding the “less-credible positive” variants; Table 1). As for other common diseases, there is increasing interest in the effects of lower-frequency SNPs and rare variants, types of genetic variation other than SNPs, and the effects of possible gene–environment and gene–gene interactions. Further, advances in technology enable the capture of large amounts of data on types of biological variations, in addition to genomic, including epigenetic, proteomic, transcriptomic, and metabolomic variations. These developments make this field synopsis the beginning of a work in progress, and we plan regular searches of the literature using the methodology described here and will continuously update the website when new data are available (http://www.chs.med.ed.ac.uk/CRCgene/).
There is an overlap of some of these loci and variants with the loci of genetic risk factors for other common complex diseases and disease traits (135). In particular, several of these genes show genome-wide associations (P < 5 × 10–8; replicated in at least one other study) with other cancers: breast cancer (CCND1), prostate cancer (8q24), and bladder cancer (GSTM1); or other diseases or disease traits: mean corpuscular hemoglobin concentration and alanine aminotransferase levels (ALDH2), ulcerative colitis (CDH1), diastolic blood pressure (CYP1A1), CRP (IL6), serum matrix metalloproteinase (MMP1 and MMP3), systolic blood pressure, and plasma homocysteine (MTHFR), Crohn’s disease and inflammatory bowel disease (NOD2). The pleiotropic associations with 8q24, MTHFR, and ALDH2 are with the same SNP that has been reported to have an association with colorectal cancer. These genome-wide associations are listed at http://www.genome.gov/gwastudies/, but because the causal genetic variant is not known in most reports, there may be some instances of wrong assignment of genes to these observed associations. Nevertheless, reports of robust associations with other diseases in 11 genes (35% of genes showing association with colorectal cancer and 17% of all meta-analyzed genes) highlight the importance of pleiotropy. This presumably reflects a common underlying pathological process underlying these conditions, for example, between (i) colorectal, breast, esophageal, bladder, and prostate cancers (CND1, MYC, GSTM1) and (ii) colorectal cancer and inflammatory bowel disease (CDH1). Other pleiotropic links are less readily explained but may yield clues to novel underlying pathological processes.
We have conducted a comprehensive exercise to capture and meta-analyze all SNP data for variants with MAFs in the range 0.01–0.49. The analysis clearly identifies 16 variants for which there is robust evidence of impact on risk of colorectal cancer; 23 variants for which further evidence through international collaboration should be generated; and 20 variants for which the overall evidence does not support association and on which further research is not warranted. With increasing availability of data from multiple SNPs, it is clear that studies to test associations must achieve very high levels of statistical stringency. It has been suggested that even for candidate SNPs, that statistical support should reach genome-wide thresholds. Nonetheless, the analysis here provides a resource for mining available data and puts into context the sample sizes required for the identification of true associations. This study highlights a number of SNP associations that could be incorporated into genetic risk–prediction algorithms as further risk factors are identified and highlights the loci at which further research effort should be targeted. The data are all lodged on the CRCgene database (http://www.chs.med.ed.ac.uk/CRCgene/).
Funding
Cancer Research, UK (C348/A3758 and A8896, C48/A6361); Medical Research Council (G0000657-53203); Scottish Executive Chief Scientist’s Office (K/OPR/2/2/D333, CZB/4/449); and a Centre Grant from CORE as part of the Digestive Cancer Campaign; Cancer Research UK Fellowship (C31250/A10107 to ET); Canadian Institutes of Health Research (CIHR) Team in Interdisciplinary Research on Colorectal Cancer (CTP-79845); a CIHR pilot project grant in colorectal cancer screening (200509CCS-152119-CCS-CECA-102806); the Cancer Risk Evaluation (CaRE) Program Grant from the Canadian Cancer Society Research Institute (18001).
References
136.
Notes
JL holds a Tier 1 Canada Research Chair in Human Genome Epidemiology.
The study sponsors had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
JL and HC conceived the study; JL, HC, ET, ZM, and SH designed it; ET, ZM, HC, and JL wrote the article with input from other authors; ET and ZM undertook data manipulations and statistical analysis; GCdLA and JG undertook the literature review; ET, ZM, SH, GCdLA, JG, VT, IK, MT, AD, LZ, DL, HEB, and SHR coordinated and/or undertook related abstraction, handling, and curation of the data; SMF, AT, and ZM provided the GWAS data for Scotland and Canada; SMF, IR, AT, and MGD provided consultation in their areas of expertise. JL, HC, MD, and ET obtained funding for the study.
The team in Edinburgh would like to thank Mrs Gisela Johnstone, Stephanie Scott, and Rosa Bisset for their administrative support and Mr Colin Pride for his IT support. The team in Ottawa would like to thank Dr Philip Ryan (University of Adelaide) for providing information that clarified the data published by Butler et al. (136).


